Figure 2.
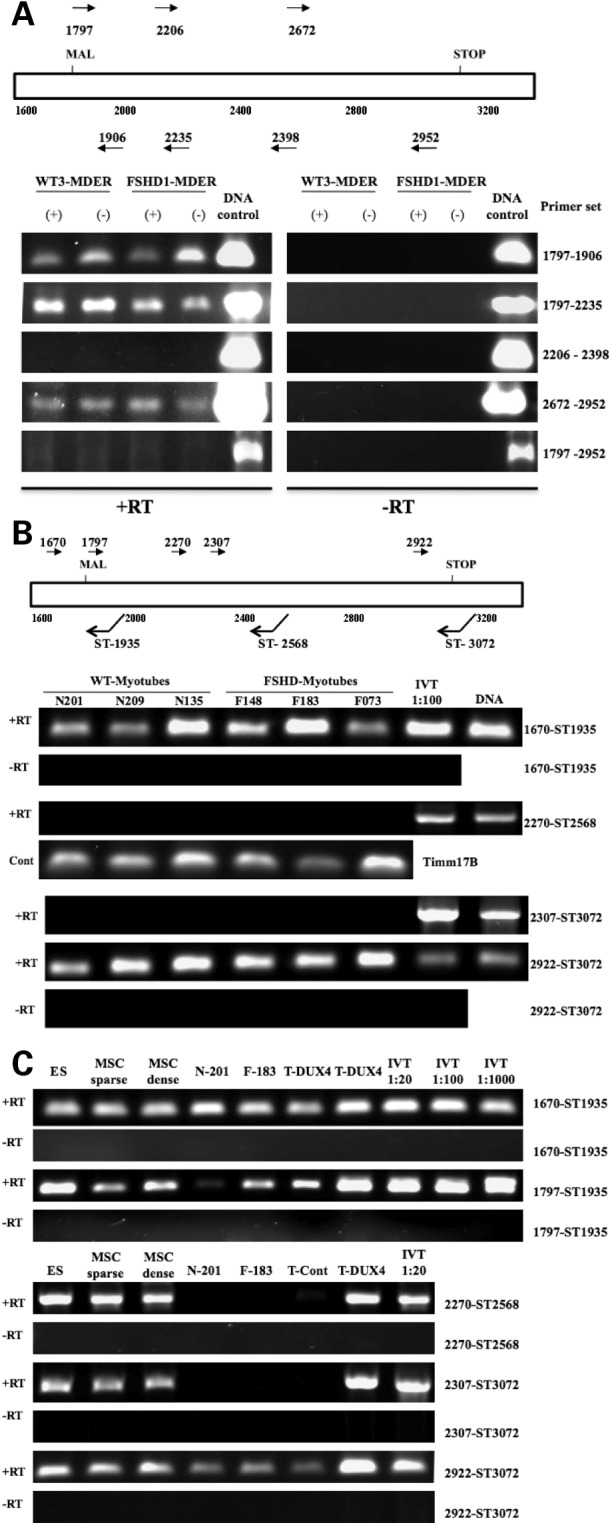
Discontinuous regions of DUX4 transcripts in wild-type and FSHD fibroblasts. (A) RT–PCR of random hexamer primed total RNA from fibroblasts, wild-type or FSHD-derived, amplified with primers for the 5-prime (1797–1906; 1797–2235), the central portion (2270–2398), the 3-prime end (2672–2970) and the full-length DUX4 (1797–2970). The fibroblasts were transduced with a retrovirus expressing MyoD-ER and assayed under differentiation conditions (DMEM with 1 µg/ml insulin and transferrin for 96 h) either without beta-estadiol (−) or with beta-estradiol (+) to induce MyoD activity; however, no consistent differences were noted in RNA obtained from MyoD induced and non-induced cells. The presence of amplification products from DNA controls shows that all primer sets can amplify the sequence from DUX4 cDNA templates. (B) Strand-specific RT–PCR amplification products using total RNA from control and FSHD muscle cells. Primer pairs are designed to amplify the 5-prime (1670-ST1936), middle (2270-ST2568) and 3-prime (2922-ST3072) regions of DUX4. IVT is a dilution of RNA from an in vitro transcribed DUX4 cDNA to demonstrate that the RT reaction and PCR can amplify the central portion of DUX4 from an RNA template. (C) Strand-specific RT–PCR amplification products using total RNA from human embryonic stem cells (ES), mesenchymal stem cells (MSC) and muscle cells from control (N-201) and FSHD (F-183) individuals. In contrast to the wild-type and FSHD muscle cells, the central portion of DUX4 can be amplified from the ES and MSC, but not from wild-type or FSHD muscle cells. Template controls include: IVT, dilutions of RNA from in vitro transcribed DUX4 cDNA; T-DUX4 and T-Control, RNA from mouse C2C12 muscle cells transfected with a DUX4 expression vector and the empty expression vector, respectively. In both (B) and (C), three independent wild-type and FSHD-derived myoblasts cultures were grown to confluence and induced to differentiate for 96 h. Similar results were obtained from undifferentiated wild-type and FSHD myoblasts (data not shown). Note that the size of the PCR product can be calculated from the position of the PCR primers.
