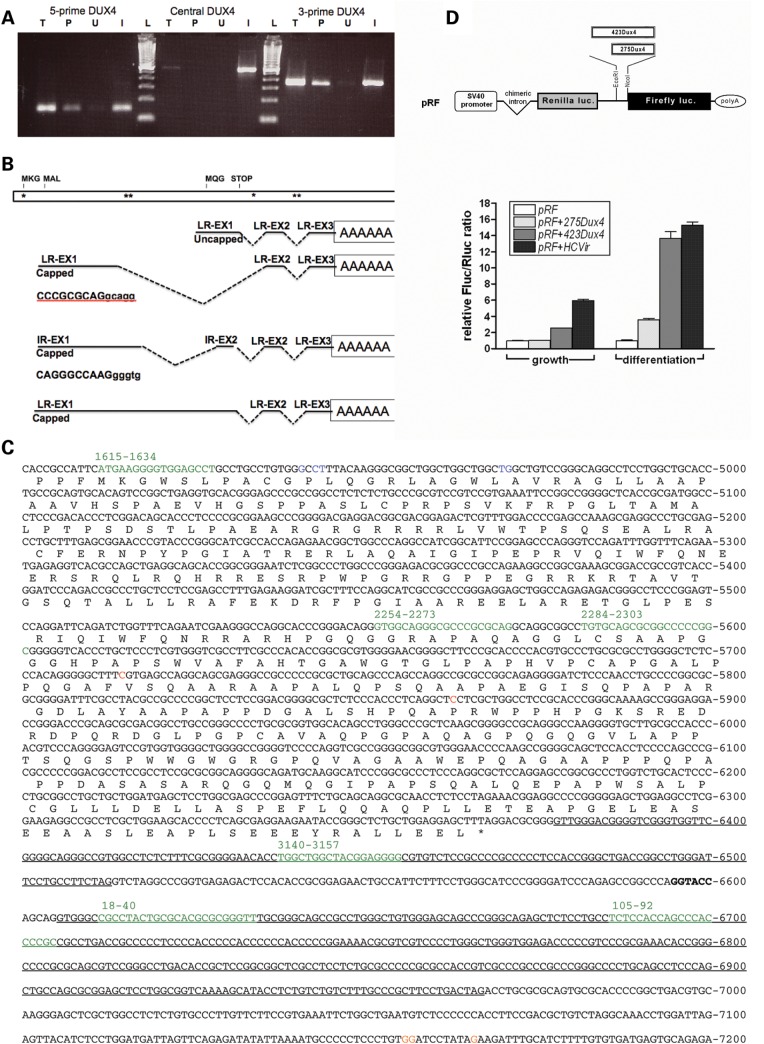
Figure 7.
Five-prime and 3-prime polyadenylated transcripts with an IRES-like element upstream of the MQG ORF. (A) RT–PCR on random primed RNA from FSHD muscle cells using primers to three regions of the DUX4 transcript (5-prime, Central and the 3-prime region of the MQG ORF) on total RNA (T), the poly-adenylated fraction that binds oligo-dT (P), or the unbound fraction that does not bind oligo-dT (U). Primers used were: 5-prime, 1707 and 1906; central, 2307and 2815; 3-prime, 6315 and 7074. In vitro transcribed full-length DUX4 RNA was used as a positive control for the RT reaction (I). 100 bp ladder (L) with 100 bp as lowest band. (B) A schematic of the DUX4 region with representations of the transcripts identified by a combination of 5-prime RACE and 3-prime RACE on the poly-A fraction and location of miRNA-like fragments indicated by asterisks. Top schematic shows locations of potential translation start codons (MKG, MAL and MQG) and stop codon (STOP); asterisks indicate locations of miRNA-like fragments; LR-EX1, cloned sequence matches last repeat-ExonI; LR-EX2, last repeat Exon 2; LR EX3, last repeat Exon 3; IR-EX1, cloned sequence does not match either first or last repeat (Supplementary Material, Fig. S4); IR-EX2, in this case cloned sequence does match LR-EX2 but the tandem repeat indicates it comes from an internal repeat. (C) Sequence of the DUX4 ORF and pLAM region showing the locations of the capped 5-prime ends (Blue, positions 4941–4944 and 4970–4971) and uncapped 5-prime ends (Red, positions 5715 and 5863), representing sites of transcription initiation and RNA cleavage, respectively. The polyadenylation sites are indicated in Orange (postions 7155–7156 and 7166); introns are underlined; miRNA-sized fragments are shown in green (note that the last partial D4Z4 unit is between the last full D4Z4 unit and the pLAM sequence (Fig. 1), and therefore, the last two miRNA-sized fragments are also present in the beginning of the D4Z4 repeat). (D) The construction of the dual cistronic pRF backbone is detailed in (33). The locations of SV40 promoter and chimeric intron are indicated, and Poly-A is the SV40 polyadenylation signal. Inserting test sequences between the EcoRI and NcoI sites of pRF created the constructs pRF + 423DUX4, pRF + 275DUX4. pRF + HCVir was created by inserting the previously characterized IRES element from the Hepatitis C virus as a positive control, and its IRES activity has been previously described (34). An empty pRF plasmid without insert was used as a negative control. Each of the constructs was transfected into mouse myoblast C2C12 cells as two sets of triplicates. Twenty-four hours post-transfection, one triplicate set of cells, designated as ‘growth,’ was harvested and their lysates assayed for FLuc and RLuc activities as described in Materials and Methods. The remaining set was switched to ‘differentiation’ media and assayed for luciferase activities 48 h post-transfection. FLuc activity was normalized to RLuc and plotted as the mean ± SD relative to the empty plasmid pRF.