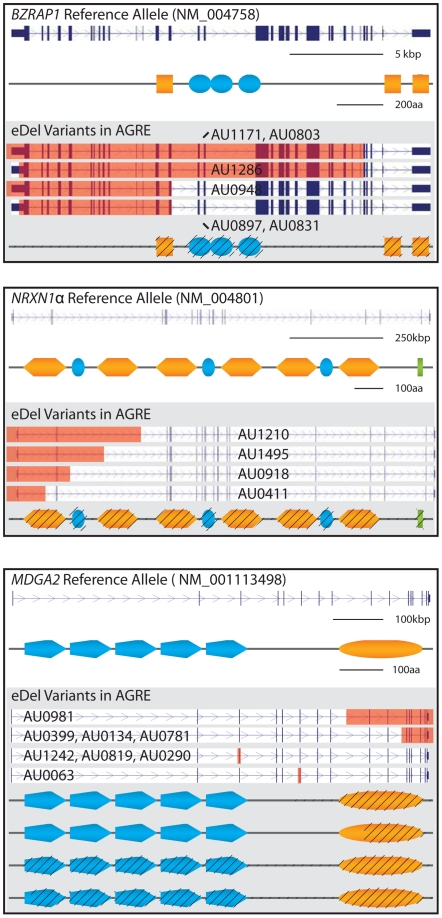
Figure 2. Rare exonic deletions (eDels) in NRXN1 and novel candidate genes alter predicted protein structures.
For each of BZRAP1 (a) NRXN1 (b) and MDGA2 (c) reference loci and encoded proteins (top) are contrasted against mutant loci and corresponding proteins (bottom; grey shading). Unique genomic deletions and corresponding protein truncations are highlighted in red and with black hatching, respectively. Schematized protein domains genes are as follows: BZRAP1—Src homology-3 (orange square), Fibronectin, type III (blue oval); NRXN1—Laminin G (orange hexagon), EGF-like (blue oval), 4.1 binding motif (green rectangle); MDGA2—IG-like domains (blue pentagon), MAM aka Meprin/A5-protein/PTPmu (blue oval).