Abstract
Nitrogenase is an essential metalloenzyme that catalyzes the biological conversion of dinitrogen (N2) to ammonia (NH3). The vanadium (V)-nitrogenase is very similar to the “conventional” molybdenum (Mo)-nitrogenase, yet it holds unique properties of its own that may provide useful insights into the general mechanism of nitrogenase catalysis. So far, characterization of the vanadium iron (VFe) protein of Azotobacter vinelandii V-nitrogenase has been focused on 2 incomplete forms of this protein: αβ2 and α2β2, both of which contain the small δ-subunit in minor amounts. Although these studies provided important information about the V-dependent nitrogenase system, they were hampered by the heterogeneity of the protein samples. Here, we report the isolation and characterization of a homogeneous, His-tagged form of VFe protein from A. vinelandii. This VFe protein has a previously-unsuspected, α2β2δ4-heterooctameric composition. Further, it contains a P-cluster that is electronically and, perhaps, structurally different from the P-cluster of molybdenum iron (MoFe) protein. More importantly, it is catalytically distinct from the MoFe protein, particularly with regard to the mechanism of H2 evolution. A detailed EPR investigation of the origins and interplays of FeV cofactor- and P-cluster-associated signals is presented herein, which lays the foundation for future kinetic and structural analysis of the VFe protein.
Nitrogenase is a complex metalloenzyme that catalyzes the biological conversion of dinitrogen (N2) to ammonia (NH3). Although the molybdenum (Mo)-dependent nitrogenase has been recognized as the “conventional” nitrogenase for the past 70 years, the vanadium (V)-nitrogenase was discovered only some 30 years ago as an “alternative” nitrogenase that is expressed under Mo-deficient conditions (1–4)*. Both nitrogenases are binary enzyme systems. The Mo-nitrogenase is composed of (i) nifH-encoded iron (Fe) protein, an α2-homodimer bridged by a [Fe4S4] cluster; and (ii) nifDK-encoded MoFe protein, an α2β2-heterotetramer containing, per αβ-dimer, a P-cluster ([Fe8S7]) that is located at the α/β-subunit interface and a FeMo cofactor (FeMoco) ([MoFe7S9X-homocitrate], where X = C, O or N) that is buried within the α-subunit (6, 7). Catalysis by this nitrogenase involves repeated association/dissociation between Fe and MoFe proteins and ATP hydrolysis-driven, interprotein electron transfer from the [Fe4S4] cluster of the Fe protein to the P-cluster, and finally, the FeMoco site of the MoFe protein, where substrate reduction occurs (1). Like its Mo-counterpart, V-nitrogenase is a 2-component system comprising (i) vnfH-encoded Fe protein and (ii) vnfDGK-encoded vanadium-iron (VFe) protein (2). The Fe protein of V-nitrogenase, a homodimer containing a [Fe4S4] cluster, is very similar to the Fe protein of Mo-nitrogenase. The VFe protein, however, differs from the α2β2-tetrameric MoFe protein in that it contains an additional δ-subunit (encoded by vnfG) along with the α- and β-subunits (encoded by vnfD and vnfK, respectively). Apart from this discrepancy, the VFe protein is also homologous to the MoFe protein, particularly with regard to the α2β2 core structure and the 2 clusters associated with the core: the P-cluster (bridged between the α- and β-subunits) and the FeV cofactor (FeVco) (located within the α-subunit). In addition, the catalytic mechanism of V-nitrogenase presumably resembles that of its Mo counterpart, which involves the interaction between Fe and VFe proteins and ATP-dependent transfer of electrons between the 2 proteins.
So far, attempts have been made to isolate the V-nitrogenase from 2 closely-related soil bacteria: Azotobacter chroococcum and Azotobacter vinelandii (8–10). The VFe protein of A. chroococcum has an optimal subunit composition of α2β2δ2 (11)†; whereas the same protein from A. vinelandii has been purified in 2 forms: αβ2 and α2β2, both of which contain the small δ-subunit in minor amounts (10). Biochemical and spectroscopic investigations of these proteins have revealed the similarities and dissimilarities between VFe and MoFe proteins and provided invaluable insights into the V-dependent nitrogenase system (2). However, the heterogeneity of the protein samples, particularly in the case of the A. vinelandii VFe protein, has hampered further progress along this line of research. Here, we report the isolation and characterization of a homogeneous, His-tagged form of VFe protein from A. vinelandii. This VFe protein has a previously-unsuspected, α2β2δ4-heterooctameric composition. Further, it contains a P-cluster that is electronically and, perhaps, structurally different from the P-cluster of MoFe protein. More importantly, it is catalytically distinct from the MoFe protein, particularly with regard to the mechanism of H2 evolution. A detailed EPR investigation of the origins and interplays of FeVco- and P-cluster-associated signals is presented herein, which lays the foundation for future kinetic and structural analysis of the VFe protein.
Results
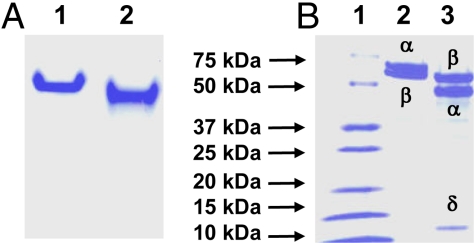
Using the fast one step purification method, up to ≈500 mg of His-tagged VFe protein was routinely purified from 250 g of cells of A. vinelandii strain YM68A. Like MoFe protein, VFe protein appears as a single band in the native PAGE (Fig. 1A), suggesting that it is a homogeneous protein species. However, VFe protein exhibits different mobility than MoFe protein in the native gel (Fig. 1A), which originates, in part, from their different subunit compositions. MoFe protein is an α2β2 tetramer comprising α (≈56 kDa) and β (≈59 kDa) subunits. In contrast, VFe protein is consisted of 3 subunits: α (≈53.9 kDa), β (≈54.1 kDa), and δ (≈13.4 kDa) (Fig. 1B); and an α/β/δ molar ratio of ≈1:1:2 is obtained from quantitative N-terminal amino acid analysis and total amino acid determination (Table 1). Moreover, gel filtration data show that VFe protein has a molecular mass of 270 kDa, which is consistent with an α2β2δ4-subunit composition of the protein. With regard to the cluster content, metal analysis reveals that VFe protein contains ≈2 mol of V and ≈34 mol of Fe per mol of protein (Table 2), which could account for the presence of 2 FeVcos and 2 P-clusters per protein molecule‡. Thus, a homogeneous species of VFe protein from A. vinelandii was obtained, which contains all 3 subunits in stoichiometric amounts. The metal content is the highest documented to date to our knowledge, suggesting the presence of a full complement of metal clusters in the protein. Moreover, the unexpected α2β2δ4-subunit composition signifies novel structural and/or functional properties of this protein.
Fig. 1.
Purification of MoFe and VFe proteins from A. vinelandii. (A) Shown is 7.5% native PAGE of MoFe and VFe proteins. Lane 1, 15 μg of purified MoFe protein; lane 2, 15 μg of purified VFe protein. (B) Shown is 4–20% gradient SDS/PAGE of MoFe and VFe proteins. Lane 1, 15 μg of protein standard; lane 2, 12.5 μg of purified MoFe protein; lane 3, 12.5 μg of purified VFe protein.
Table 1.
Subunit composition of VFe protein from A. vinelandii
| Subunit | Amount,μg | Molecular mass,g/mol | Amount,pmol | Ratio(normalized) |
|---|---|---|---|---|
| α (VnfD) | 0.9 | 53,880 | 16.7 | 1.1 |
| β (VnfK) | 0.8 | 54,100 | 14.8 | 1.0 |
| δ (VnfG) | 0.4 | 13,370 | 29.9 | 2.0 |
The amount of each subunit was determined by total amino acid sequencing by Molecular Structure Facility at the University of California, Davis. Consistent with this result, an α/β/δ ratio of 1.0:1.0:1.8 was obtained from quantitative sequencing of N-terminal amino acids by Agnes Henschen-Edman at the University of California, Irvine.
Table 2.
Metal composition of VFe protein from A. vinelandii
| V (mol V/mol protein) | Fe (mol Fe/mol protein) |
|---|---|
| 1.9 ± 0.3 | 33.5 ± 3.8 |
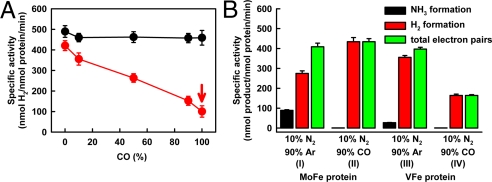
Consistent with earlier reports (2), VFe protein differs from MoFe protein in catalysis (Table 3). Compared with MoFe protein, the total electron fluxes through VFe protein under C2H2/Ar (where C2H4 and C2H6 are formed), Ar (where only H2 is formed), and N2 (where NH3 and H2 are formed) are 30%, 86%, and 81%, respectively (Table 4). As such, VFe protein is less effective in substrate reduction than MoFe protein. C2H2 appears to be a particularly poor substrate for VFe protein; however, it can be reduced to both C2H4 and C2H6 (in minor quantity). In comparison, MoFe protein reduces C2H2 with a much higher efficiency, yet C2H4 is the only product formed in this reaction. Under N2, VFe protein generates NH3 and H2 at a NH3/H2 ratio of 0.9, whereas MoFe protein forms these 2 products at a NH3/H2 ratio of 2.3 (Table 4). Clearly, there is a shift of electron flow toward H2 evolution in the reaction catalyzed by VFe protein.
Table 3.
Specific substrate-reducing activities of VFe and MoFe proteins of A. vinelandii
| Protein | nmol product/nmol protein per min |
||||
|---|---|---|---|---|---|
| C2H4 formation under C2H2 | C2H6 formation under C2H2 | NH3 formation under N2 | H2 formation under N2 | H2 formation under Ar | |
| MoFe | 483 ± 19 | 0 | 205 ± 5 | 133 ± 8 | 489 ± 34 |
| VFe | 136 ± 3 | 4 ± 1 | 111 ± 10 | 192 ± 7 | 419 ± 8 |
Activities of MoFe protein and VFe protein were determined with nifH-encoded and vnfH-encoded Fe proteins, respectively. Activity assays were performed at pH 5.0, 6.0, 7.0 and 8.0, and the optimal activities of both MoFe and VFe proteins were achieved at pH 8.0.
Table 4.
Specific substrate-reducing activities based on electron pairs of VFe and MoFe proteins of A. vinelandii
| Protein | Condition* | Electron pair appearing in product† |
Ratio of NH3/H2 | Sum of activity‡ | |||
|---|---|---|---|---|---|---|---|
| C2H4 | C2H6 | H2 | NH3 | ||||
| MoFe | C2H2/Ar | 489 ± 19 | — | — | — | — | 483 ± 19 (100) |
| Ar | — | — | 489 ± 34 | — | — | 489 ± 34 (100) | |
| N2 | — | — | 133 ± 8 | 308 ± 8 | 2.3 | 441 ± 16 (100) | |
| VFe | C2H2/Ar | 136 ± 3 | 8 ± 2 | — | — | — | 144 ± 5 (30) |
| Ar | — | — | 419 ± 8 | — | — | 419 ± 8 (86) | |
| N2 | — | — | 192 ± 7 | 167 ± 15 | 0.9 | 359 ± 22 (81) | |
*Products formed under the conditions listed are: (i) C2H4 and C2H6 (under C2H2/Ar); (ii) H2 (under Ar); and (iii) NH3 and H2 (under N2). The reactions generating these products are: (i) C2H2 + 2H+ + 2e− → C2H4 and C2H2 + 4H+ + 4e− → C2H6; (ii) 2H+ + 2e− → H2; and (iii) N2 + 8H+ + 8e− → 2NH3 + H2.
†Activity expressed as nmol electron pair appearing in product/min per nmol protein, calculated by multiplying activity in nmol product/min per nmol protein by the number of electron pairs appearing in each product, namely, 1, 2, 1 and 1.5, respectively, for C2H4, C2H6, H2 and NH3. This approach has been used previously to calculate the specific activities of nitrogenase reactions that involve the concomitant formation of multiple products (12, 13).
‡The sum of activity under each condition is calculated by adding activities, expressed in nmol electron pair/min per nmol protein, of formation of products concurrently formed in the same reaction. Thus-calculated activity represents the total electron flux per reaction, which is a better measure for activities in reactions involving multiple products. Percentages of activities of VFe protein in comparison to those of MoFe protein are given in parentheses.
The discrepancy between the catalytic properties of VFe and MoFe proteins is further demonstrated by the effect of CO, a well-established inhibitor of nitrogenase, on H2 evolution. Under Ar, H2 formation by MoFe protein is largely unaffected by the presence of CO (Fig. 2A, black line). On the contrary, the same reaction catalyzed by VFe protein is gradually suppressed by an increasing amount of CO; however, it cannot be completely inhibited, preserving an activity of ≈108 nmol/nmol protein per min even at 100% CO (Fig. 2A, red line). The same discrepancy in the effect of CO on H2 evolution is observed for MoFe and VFe proteins in the presence of 10% N2, where H2 is formed as a coproduct of N2 reduction. Formation of NH3 from N2 by either MoFe or VFe protein (Fig. 2B, I and III, black bars) is completely inhibited by the presence of 90% CO (Fig. 2B, II and IV, black bars). Consequently, the electrons associated with NH3 formation are rerouted to the formation of H2. Because CO does not affect H2 formation by MoFe protein, the rerouted electrons contribute to a net gain in H2 evolution (Fig. 2B, I and II, red bars). In the case of VFe protein, however, the inhibitory effect of CO on H2 formation (Fig. 2A) outweighs the small gain of H2 from rerouted electrons. As a result, there is a net loss in H2 evolution by VFe protein under CO (Fig. 2B, III and IV, red bars). Interestingly, the total electron flux through MoFe protein remains constant in the presence or absence of CO (Fig. 2B, I and II, green bars), whereas the total electron flux through VFe protein is significantly decreased in the presence of CO (Fig. 2B, III and IV, green bars). Taken together, these observations suggest a mechanistic difference between the 2 proteins.
Fig. 2.
Effect of CO on H2 formation by MoFe and VFe proteins. (A) Specific activity of H2 evolution with increasing amount of CO (balanced with Ar) by MoFe protein (black) and VFe protein (red). (B) Specific activities of NH3 formation (black bars), H2 formation (red bars), and total electron pairs that appear in NH3 and H2 (green bars) <10% N2/90% Ar and 10% N2/90% CO, respectively, in reactions catalyzed by MoFe protein (I and II) and VFe protein (III and IV).
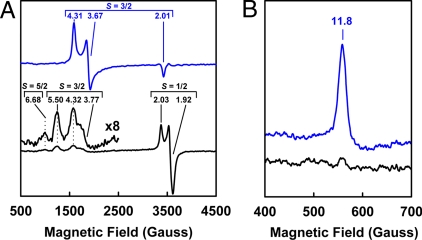
The distinct catalytic profile of VFe protein correlates further with its unique EPR features, which are different from those of the MoFe protein. In the dithionite-reduced state, the MoFe protein displays a rhombic S = 3/2 signal that originates from the FeMoco center (1); in contrast, the VFe protein exhibits 3 EPR signals, which are 1/2, 3/2 and 5/2 in terms of spin property (Fig. 3A). Although the S = 3/2 signal of VFe protein has been assigned to the active FeVco center, the S = 1/2 and 5/2 signals have not been well defined (2, 11). In the indigo disulfonate (IDS)-oxidized state, the MoFe protein exhibits a strong, g = 11.8 signal in the parallel-mode EPR that is associated with the +2 oxidation state of the P-cluster (P2+) (14, 15). Such a signal is nearly invisible (if any) in the spectrum of VFe protein (Fig. 3B). Clearly, the cluster species in VFe protein differ from those in MoFe protein in terms of electronic and, perhaps, structural properties. So far, an unambiguous assessment of the EPR properties of metal clusters in VFe protein has been precluded by the heterogeneity of protein samples. The success in obtaining a homogeneous and fully-complemented form of VFe protein allows us to visualize, within 1 protein species, all previously-reported EPR features (which spread among different forms of VFe proteins from different organisms) at higher intensity and better resolution. Such an “all-in-one” form of VFe protein presents an unprecedented opportunity for a detailed investigation of the origins and interplays of the various EPR features of VFe protein, which, in turn, permits an in-depth account for the unique properties of VFe protein from a spectroscopic perspective.
Fig. 3.
EPR properties of MoFe and VFe proteins. (A) Perpendicular-mode EPR spectra of dithionite-reduced MoFe protein (blue) and VFe protein (black). The S = 5/2, 3/2 and 1/2 signals are indicated, and the g values of each signal are shown. (B) Parallel-mode EPR spectra of IDS-oxidized MoFe protein (blue) and VFe protein (black). The P-cluster (P2+) specific, g = 11.8 signal is indicated. Spectra were collected at 50 mW and 15 K.
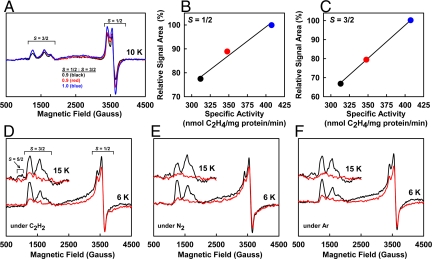
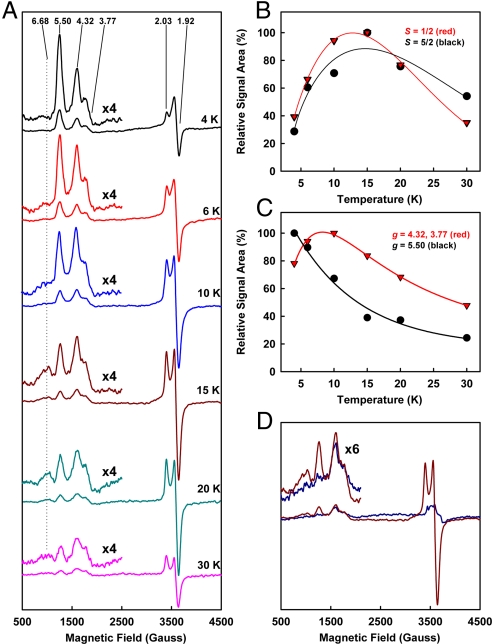
The S = 1/2 signal is perhaps the feature that best distinguishes VFe protein from MoFe protein in the dithionite-reduced state. It is an axial signal with g values of 2.03 and 1.92, and it integrates to 0.4 spin per protein (Fig. 3A). Conflicting data have been presented concerning the physiological relevance of this signal (11). Our data show that (i) the intensity of the S = 1/2 signal remains at a constant ratio to that of the FeVco-associated, S = 3/2 signal among VFe protein samples of varying activities (Fig. 4A); (ii) the magnitude of the S = 1/2 signal (Fig. 4B), like that of the S = 3/2 signal (Fig. 4C), is proportional to the specific activity of the protein; and (iii) in the presence of dithionite, MgATP and substrates, there is a concomitant decrease of the S = 1/2 and S = 3/2 signals in magnitude (Fig. 4 D–F). These observations suggest that the S = 1/2 signal is intimately associated with an active from of VFe protein, not only in the resting state, but also during substrate turnover.
Fig. 4.
Physiological relevance of the S = 1/2 signal of VFe protein. (A) Perpendicular-mode EPR spectra of dithionite-reduced VFe proteins. Shown are 3 samples of varying specific activities of C2H4 formation: 313 (black), 348 (red), and 407 (blue) nmol/mg protein per min. The ratio of intensity between the S = 1/2 and S = 3/2 signals of these VFe protein samples (as shown) remains nearly constant. Spectra were collected at 6 K. (B and C) Relative intensities of the S = 1/2 signals (B) and the FeVco-associated, S = 3/2 signals (C) plotted against the specific activities of the 3 VFe protein samples (represented by black, red, and blue circles, as in A). (D–F) Changes of EPR spectra in the presence of C2H2 (D), N2 (E), and Ar (F) under turnover conditions. Shown are spectra of dithionite-reduced VFe proteins in resting (black) and turnover (red) states. Concomitant with the attenuation of the FeVco-associated S = 3/2 signal upon turnover, S = 1/2 signal is diminished in intensity, most notably at 6 K. The S = 5/2 signal is also reduced in magnitude during turnover; however, such a change is best visualized at 15 K (Inset).
The origin of this S = 1/2 signal has also remained a topic of debate. Earlier Mössbauer experiments suggest that the S = 1/2 signal arises from FeVco (16). However, such an S = 1/2 signal has been observed in a FeVco-deficient, yet P-cluster-containing form of VFe protein (generated by deletion of nifB, the starting point of FeVco biogenesis) (17). This observation suggests that the P-cluster, instead of the FeVco, gives rise to this signal. In this scenario, the P-cluster center of the VFe protein would be different from the “standard” P-cluster of the MoFe protein in spectroscopic properties, because (i) the P-cluster of MoFe protein does not show an S = 1/2 signal in the dithionite-reduced state; and (ii) the P-cluster (P2+)-associated signal of the IDS-oxidized MoFe protein is not clearly visible in the spectrum of the VFe protein (Fig. 3B). Interestingly, previous oxidative titrations show that the +1 oxidation state of the P-cluster (P1+) in partially-oxidized MoFe protein displays an S = 1/2 signal with g values of 2.06 and 1.95 (15). The presence of an analogous S = 1/2 signal in the dithionite-reduced VFe protein, therefore, suggests that the P-cluster species of the VFe protein may exist in a more oxidized state than those in the MoFe protein. One possible explanation for such a discrepancy is that the P-cluster in VFe protein is structurally distinct from its counterpart in MoFe protein. In support of this argument, the extended x-ray absorption fine structure (EXAFS) spectrum of the FeVco-deficient VFe protein closely resembles that of the FeMoco-deficient ΔnifH MoFe protein, which contains a pair of [Fe4S4]-like clusters in place of the [Fe8S7] cluster (17).
The S = 5/2 signal of VFe protein (18) has a g value of 6.68 (Fig. 3A) and appears to be closely associated with the S = 1/2 signal, because (i) it has a similar temperature-dependent pattern as the S = 1/2 signal (Fig. 5B); and (ii) it disappears concurrently with the S = 1/2 signal upon IDS oxidation (Fig. 5D). Additionally, like the S = 1/2 signal, an analogous S = 5/2 signal has also been observed at g = 6.70 in the partially-oxidized MoFe protein and assigned to the P-cluster in the +1 oxidation state (P1+) (15). Thus, the P-cluster of VFe protein could be the common origin of both S = 1/2 and S = 5/2 signals.
Fig. 5.
Temperature-dependent and redox-associated changes of EPR signals of VFe protein. (A) Variation of the EPR signals of dithionite-reduced VFe protein with increasing temperature. The S = 5/2 and 3/2 regions are enlarged and shown in the Insets. The g values of all signals are indicated. (B) Similarity in temperature dependency between the S = 1/2 (red) and 5/2 (black) signals. (C) Difference in temperature dependency between the combined feature at g = 4.32 and 3.77 (red) and the feature at g = 5.50 (black) of the S = 3/2 signal. (D) EPR spectra of VFe protein in dithionite-reduced (brown) and IDS-oxidized (blue) states. Upon IDS oxidation, the S = 5/2 and 1/2 signals and the g = 5.50 feature of S = 3/2 signal decrease in intensity; whereas the g = 4.32 and 3.77 features of S = 3/2 signal remain largely unchanged.
The S = 3/2 signal is the most complex among all EPR features of VFe protein (2, 11), exhibiting g values of 5.50, 4.32, and 3.77 (Fig. 3A). Like the S = 3/2 signal of the MoFe protein, this signal has been assigned to the substrate reducing site, i.e., the FeVco center of the VFe protein (2, 11). The S = 3/2 signal of the VFe protein also behaves like that of the MoFe protein in that (i) its intensity is linearly correlated with the activity (Fig. 4C); and (ii) it is largely attenuated upon turnover (Fig. 4 D–F). Apparently, there is a certain degree of similarity between the 2 cofactors (i.e., FeVco and FeMoco) that give rise to these S = 3/2 signals. However, earlier magnetic circular dichroism (MCD) studies indicate that the electronic and magnetic properties of FeVco are quite distinct from those of FeMoco (19). Consistent with this observation, the S = 3/2 signal of VFe protein is more complex in shape than that of the MoFe protein, leading to the hypothesis that this signal is a mixture of several S = 3/2 species (2, 11). Indeed, the 3 features of S = 3/2 signal (at g = 5.50, 4.32, and 3.77) do not behave in synchrony, because (i) the g = 5.50 feature shows a different temperature dependency (Fig. 5C, black line) than that of the combined feature at g = 4.32 and 3.77 features (Fig. 5C, red line); and (ii) upon IDS oxidation, the g = 4.32 and 3.77 features remain largely unchanged, whereas the g = 5.50 feature disappears completely (Fig. 5D). These observations suggest that the g = 5.50 feature is a different S = 3/2 species than that associated with the g = 4.32 and 3.77 features.
Discussion
The α2β2δ4 composition of the VFe protein is somewhat surprising, given the fact that vnfD, vnfG, and vnfK genes are present in a 1:1:1 ratio within 1 operon of the chromosome (2). Explanation for such a subunit composition may be supplied by an incomplete, αβ2-trimeric form of FeVco-deficient VFe protein (17) and the αβ2-trimeric species that is likely a breakdown product generated by the prolonged purification procedures of nontagged VFe protein (10). Apparently, the δ-subunit is required for the stabilization of the α/β-subunit interface, because the absence of this subunit results in the dissociation of 1 αβ-dimer and the loss of 1 α-subunit. Should this be the case, the 4 δ-subunits may exist as 2 pairs of δ2-dimers, each “locking” 1 αβ-dimer in the α2β2δ4 structure (Fig. S1A). Apart from stabilizing the α/β-subunit interface, the δ-subunit may also be involved in delivering FeVco to its destined location in the α-subunit, because the δ-subunit is absent from the VFe protein structure when FeVco is not around to be transported (e.g., the FeVco-deficient VFe protein). An analogous FeMoco transporter (encoded by nifY) has been identified for the MoFe protein of Klebsiella pneumonia; only in this case, the transporter protein dissociates from the α2β2 structure once its mission is completed (20). The permanent association of the δ-subunit as a δ2-dimer at the α/β interface of VFe protein may present a challenge to an unobstructed interaction between VFe and Fe proteins, because the Fe protein is another homodimer that interacts with both α- and β-subunits during catalysis. However, the δ2-dimer (26 kDa) is likely too small in size to interfere with this process; alternatively, it may have a different binding site than the Fe protein; finally, it could undergo structural rearrangements that accommodate the productive docking of Fe protein at the α/β interface of VFe protein (Fig. S1A).
Evolution of H2 by VFe protein clearly differs from that by MoFe protein. Based on the pattern of CO inhibition, there seems to be 2 different mechanisms of H2 formation by V-nitrogenase: one is suppressed by CO, the other is not (Fig. 2). In contrast, H2 evolution by Mo-nitrogenase is largely unaffected by the presence of CO (Fig. 2). Thus, the VFe protein, or, more specifically, its FeVco site may provide 2 sites for H2 evolution, and CO particularly targets one of them. Alternatively, FeVco may have only 1 site for H2 formation; however, if VFe protein follows a reaction mechanism that is similar to the Lowe–Thorneley model of MoFe protein, where electrons and H+ are added stepwise to the cofactor site during enzymatic turnover, H2 can be released at different oxidation states of the enzyme depending on how many electrons flow through the protein (Fig. S1B). Should this be the case, H2 evolution can be arbitrarily divided into 2 categories: “low-flux H2” and “high-flux H2” (Fig. 6B). In the case of VFe protein, CO likely blocks the electron flow before H2 can be evolved as a coproduct of N2 binding/reduction (Fig. S1B, red box). Consequently, only low-flux H2 is generated, which corresponds to the unsuppressed activity of H2 evolution in the presence of 100% of CO (Fig. 2A, arrow). In the case of MoFe protein, however, CO may block the electron flow at a later stage (Fig. S1B, green box), preventing the release of NH3, yet leaving the evolution of H2 intact. A third account for the differential effect of CO on H2 evolution by MoFe and VFe proteins is that the addition of CO raises the resting E0′ of the cofactors in the respective proteins without removing any electrons. The resting E0′ of the MoFe protein could be more negative than that of the VFe protein. Thus, an increase in E0′ upon the addition of CO may render the MoFe protein incapable of reducing N2, yet still fully capable of producing H2; whereas in the case of the VFe protein, which starts from a more positive E0′, the addition of CO may inhibit N2 reduction and diminish H2 production at the same time.
The unique substrate-reducing activities of VFe protein may very well be associated with the presence of unique cluster species in the protein. The P-cluster in VFe protein is likely more oxidized and, perhaps, more “open” in structure, than the “standard” P-cluster in MoFe protein. This possibility is supported by the observation that the P-cluster in MoFe protein can undergo a two-electron oxidation process that concurrently opens up half of the [Fe8S7] cage (6). Further, in the resting state, the P-cluster in VFe protein gives rise to signals that are analogous to its MoFe protein counterpart in the turnover state (18), suggesting a somewhat interconvertible nature of these P-clusters in oxidation state and/or structure. More importantly, this observation points to a plausible redox and/or structural change of the P-cluster that may be mechanistically-relevant to catalysis. With regard to FeVco, earlier EXAFS analyses show that the Fe–Fe distances of the isolated FeVco are similar to those of the FeMoco, whereas the Fe–V distance is significantly longer than the Fe–Mo distance (21). However, despite the similarities between FeVco and FeMoco in the overall structure, MoFe protein heterologously reconstituted with isolated FeVco is only capable of reducing C2H2 and H+ at low efficiencies, and it cannot reduce N2 (22). It is possible that, in the previously-isolated FeVco, the V atom may be loosely attached to the core (which explains the long Fe–V distance) and, consequently, it either dissociates from the core structure easily or is “stuck” in an unproductive conformation (which accounts for its low/missing activity in reconstitution). Indirectly, these results suggest the impact of heterometal (V in this case) on nitrogenase activity. Further, V may convey some very distinctive magnetic and electronic properties to FeVco, which is attested to by MCD, Mössbauer, and EPR analyses (2, 11). This unique cofactor, along with the distinctive P-cluster of VFe protein, will remain the focal point of structural–functional investigations of V-dependent nitrogenase.
Materials and Methods
Unless noted otherwise, all chemicals and reagents were obtained from Fisher Scientific or Sigma–Aldrich. Cell growth, protein purification, protein characterization, activity assays, metal analysis, and EPR spectroscopy were performed as described (23). See SI Text for more information on these procedures.
Construction of A. vinelandii Strain YM68A.
A. vinelandii strain YM68A was constructed on the basis of CA11.6, a tungsten (W)-resistant A. vinelandii strain containing deletions of nifHDK on the chromosome (24). Using a previously-described procedure (25), a sequence encoding 8 histidines was inserted at the 5′ end of the vnfK gene and introduced into the genome of CA11.6. The resulting strain is designated YM68A, which expresses VFe protein with a polyhistidine tag at the N terminus of the β-subunit.
Supplementary Material
Acknowledgments.
This work was supported by Herman Frasch Foundation Grant 617-HF07 (to M.W.R.) and National Institutes of Health Grant GM-67626 (to M.W.R.).
Footnotes
The authors declare no conflict of interest.
Four classes of nitrogenase have been identified. They are the Mo nitrogenase, the V nitrogenase, the Fe-only nitrogenase, and the nitrogenase from Streptomyces thermoautotrophicus (1, 2, 5). The major distinctive feature of the first 3 classes of nitrogenase, which are otherwise similar, is the heterometal atom at the active site of cofactor (Mo, V, and Fe, respectively). The 4th nitrogenase is superoxide-dependent and quite different from the other nitrogenase classes (5).
The optimal subunit composition of A. chroococcum VFe protein was likely proposed on the basis of the genetic sequence that encodes the protein (i.e., vnfDGK), because a molecular mass of 210 kDa was reported and assigned to the presence of 2 α-subunits (50 kDa) and 2 β-subunits (55 kDa) in this protein.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904408106/DCSupplemental.
The assignment of clusters is based on the assumption that the P-cluster and FeVco in VFe protein have nearly identical metal compositions as those of their respective counterparts in MoFe protein (except for the presence of a different heterometal at the active site).
References
- 1.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Eady RR. Structure–function relationships of alternative nitrogenases. Chem Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 3.Bishop PE, Jarlenski DM, Hetherington DR. Expression of an alternative nitrogen fixation system in Azotobacter vinelandii. J Bacteriol. 1982;150:1244–1251. doi: 10.1128/jb.150.3.1244-1251.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop PE, Jarlenski DM, Hetherington DR. Evidence for an alternative nitrogen fixation system in Azotobacter vinelandii. Proc Natl Acad Sci USA. 1980;77:7342–7346. doi: 10.1073/pnas.77.12.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribbe M, Gadkari D, Meyer O. N2 fixation by Streptomyces thermoautotrophicus involves a molybdenum-dinitrogenase and a manganese-superoxide oxidoreductase that couple N2 reduction to the oxidation of superoxide produced from O2 by a molybdenum-CO dehydrogenase. J Biol Chem. 1997;272:26627–26633. doi: 10.1074/jbc.272.42.26627. [DOI] [PubMed] [Google Scholar]
- 6.Peters JW, et al. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry. 1997;36:1181–1187. doi: 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- 7.Einsle O, et al. Nitrogenase MoFe-protein at 1.16-Å resolution: A central ligand in the FeMo-cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 8.Robson RI, et al. The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme. Nature. 1986;322:388–390. [Google Scholar]
- 9.Hales BJ, Case EE, Morningstar JE, Dzeda MF, Mauterer LA. Isolation of a new vanadium-containing nitrogenase from Azotobacter vinelandii. Biochemistry. 1986;25:7251–7255. doi: 10.1021/bi00371a001. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard CZ, Hales BJ. Isolation of two forms of the nitrogenase VFe protein from Azotobacter vinelandii. Biochemistry. 1996;35:472–478. doi: 10.1021/bi951429j. [DOI] [PubMed] [Google Scholar]
- 11.Eady RR. Current status of structure function relationships of vanadium nitrogenase. Coordin Chem Rev. 2003;237:23–30. [Google Scholar]
- 12.Fisher K, Dilworth MJ, Kim C-H, Newton WE. Azotobacter vinelandii nitrogenases with substitutions in the FeMo-cofactor environment of the MoFe protein: Effects of acetylene or ethylene on interactions with H+, HCN, and CN−. Biochemistry. 2000;39:10855–10865. doi: 10.1021/bi0001628. [DOI] [PubMed] [Google Scholar]
- 13.Fay AW, Hu Y, Schmid B, Ribbe MW. Molecular insights into nitrogenase FeMoco insertion: The role of His-274 and His-451 of MoFe protein α-subunit. J Inorg Biochem. 2007;101:1630–1641. doi: 10.1016/j.jinorgbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierik AJ, Wassink H, Haaker H, Hagen WR. Redox properties and EPR spectroscopy of the P clusters of Azotobacter vinelandii MoFe protein. Eur J Biochem. 1993;212:51–61. doi: 10.1111/j.1432-1033.1993.tb17632.x. [DOI] [PubMed] [Google Scholar]
- 15.Tittsworth RC, Hales BJ. Detection of EPR signals assigned to the 1-equiv-oxidized P-clusters of the nitrogenase MoFe-protein from Azotobacter vinelandii. J Am Chem Soc. 1993;115:9763–9767. [Google Scholar]
- 16.Ravi N, Moore V, Lloyd SG, Hales BJ, Huynh BH. Mössbauer characterization of the metal clusters in Azotobacter vinelandii nitrogenase VFe protein. J Biol Chem. 1994;269:20920–20924. [PubMed] [Google Scholar]
- 17.Hu Y, et al. Nitrogenase reactivity with P-cluster variants. Proc Natl Acad Sci USA. 2005;102:13825–13830. doi: 10.1073/pnas.0506967102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tittsworth RC, Hales BJ. Oxidative titration of the nitrogenase VFe protein from Azotobacter vinelandii: An example of redox-gated electron flow. Biochemistry. 1996;35:479–487. doi: 10.1021/bi951430i. [DOI] [PubMed] [Google Scholar]
- 19.Morningstar JE, Johnson MK, Case EE, Hales BJ. Characterization of the metal clusters in the nitrogenase molybdenum-iron and vanadium-iron proteins of Azotobacter vinelandii using magnetic circular dichroism spectroscopy. Biochemistry. 1987;26:1795–1800. doi: 10.1021/bi00381a001. [DOI] [PubMed] [Google Scholar]
- 20.Homer MJ, Paustian TD, Shah VK, Roberts GP. The nifY product of Klebsiella pneumoniae is associated with apodinitrogenase and dissociates upon activation with the iron-molybdenum cofactor. J Bacteriol. 1993;175:4907–4910. doi: 10.1128/jb.175.15.4907-4910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey I, et al. Iron K-edge X-ray-absorption spectroscopy of the iron-vanadium cofactor of the vanadium nitrogenase from Azotobacter chroococcum. Biochem J. 1990;266:929–931. [PMC free article] [PubMed] [Google Scholar]
- 22.Smith BE, Eady RR, Lowe DJ, Gormal C. The vanadium-iron protein of vanadium nitrogenase from Azotobacter chroococcum contains an iron-vanadium cofactor. Biochem J. 1988;250:299–302. doi: 10.1042/bj2500299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Fay AW, Dos Santos PC, Naderi F, Ribbe MW. Characterization of Azotobacter vinelandii nifZ deletion strains: Indication of stepwise MoFe protein assembly. J Biol Chem. 2004;279:54963–54971. doi: 10.1074/jbc.M408983200. [DOI] [PubMed] [Google Scholar]
- 24.Chisnell JR, Premakumar R, Bishop PE. Purification of a second alternative nitrogenase from a nifHDK deletion strain of Azotobacter vinelandii. J Bacteriol. 1988;170:27–33. doi: 10.1128/jb.170.1.27-33.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christiansen J, Goodwin PJ, Lanzilotta WN, Seefeldt LC, Dean DR. Catalytic and biophysical properties of a nitrogenase apo-MoFe protein produced by a nifB-deletion mutant of. Azotobacter vinelandii. Biochemistry. 1998;36:12611–12623. doi: 10.1021/bi981165b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.