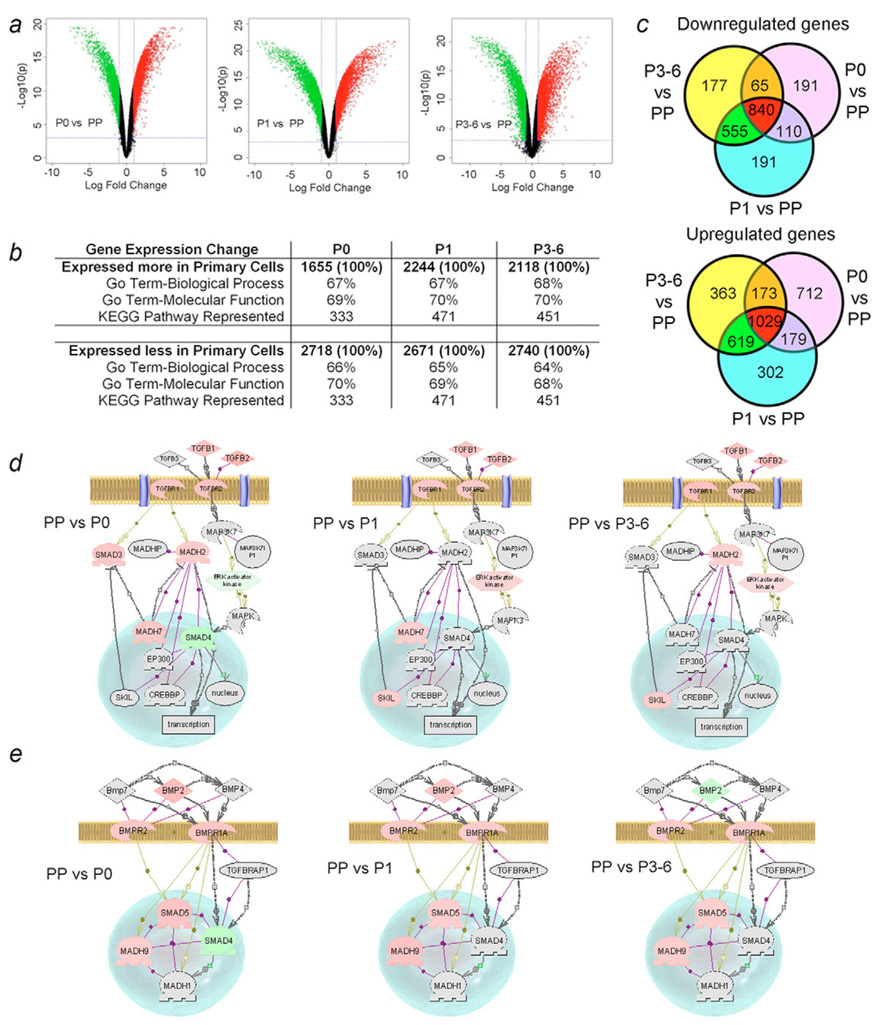
Fig. 7.
Microarray analysis of gene expression in fetal liver cells. (a) Volcano plots of gene expression in freshly isolated cells (PP) vs P0, P1 or P3-P6 cultures. Upper right and left regions show >twofold and statistically significant differences in gene expression (green=down, red=up). (b) Overall distribution of differentially expressed genes and fractions representing two gene ontology groups and KEGG pathways. (c) Identification of genes expressed in various cell populations. The data were from total gene sequences considered to have been expressed, range, 44,256 to 50,319. Compared with PP cells, 191 genes were uniquely downregulated in P0 and P1 and 177 genes were uniquely downregulated in P3-P6. By contrast, 712, 302 and 363 genes were upregulated in P0, P1 and P3-P6, respectively. Changes in TGFβ signaling (d) and BMP signaling (e) in cultured cells compared with PP cells. Genes with higher expression in PP are in pink, those expressed less well in PP are in green, and those not represented are in grey. In PP cells, Smad2 (MADH2) and Smad3 expression was consistent with their positive roles in endoderm development and hepatic differentiation, while Smad7 (MADH7, the negative regulator of Smad2 and Smad3) was expressed simultaneously, and Smad4 and ERK activator kinase were less active. By contrast, during subsequent cell culture, Smad7 inhibition was removed, along with downregulation of ERK activator kinase and Ski-like (SKIL), which inhibit the Smad signaling regulating endodermal differentiation. Analysis of BMP signaling (e) indicated that Smad4 was expressed more highly in P0 cells and BMP2 was expressed more highly in P3-P6 cells. Depending on the context of Smad4 activity, BMP2 signaling can favor mesoderm or endoderm induction. Taken together, perturbations in TGFβ and BMP signaling were in agreement with the fetal cell phenotypes observed.