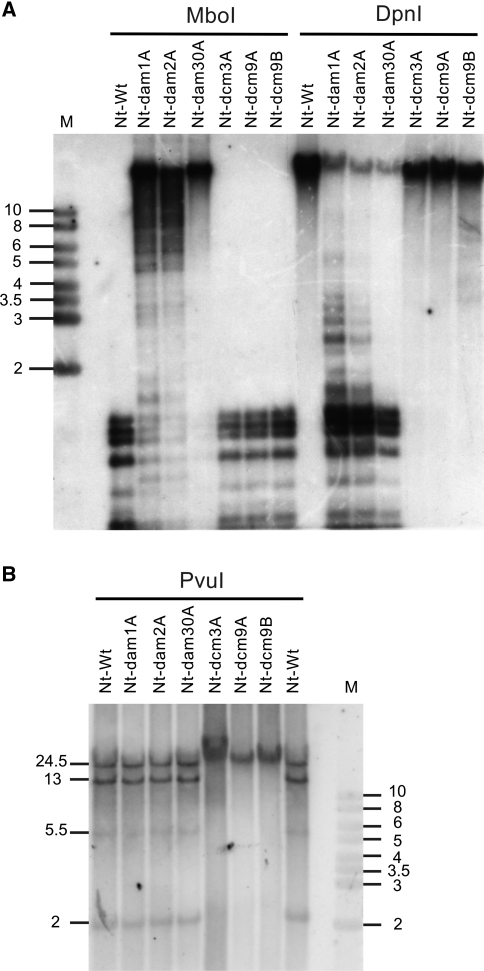
Fig. 2.
Analysis of DNA methylation in the plastid genome of Nt-dam and Nt-dcm plants. The same plant lines as in Fig. 1c were analyzed. a Test for adenine methylation with two isoschizomeric restriction enzymes differing in their sensitivity to adenine methylation in their recognition sequence (GATC). Whereas MboI cleaves the unmethylated sequence GATC but not the methylated sequence GmetATC, DpnI cleavage is dependent on Dam methylation and the unmethylated sequence is not cut (http://rebase.neb.com/rebase/rebase.html). The blot was hybridized to a 7.2 kb SalI/StuI fragment derived from the trnR(UCU)-rps2 region of the tobacco plastid genome (Shinozaki et al. 1986). Complete digestion of the plastid DNA with MboI in the wild-type (Nt-Wt) and all Nt-dcm lines indicates absence of Dam methylation. Strongly inhibited MboI cleavage in all Nt-dam lines suggests intense Dam methylation. Efficient cleavage of the plastid genome in all Nt-dam lines with the methylation-dependent restriction enzyme DpnI confirms efficient adenine methylation of GATC sequences in the Nt-dam transplastomic plants, but not the wild-type and the Nt-dcm plants. Minor differences between transplastomic lines obtained from the same construct are due to unequal loading. b Test for cytosine methylation by DNA digestion with the methylation-sensitive restriction endonuclease PvuI (Scharnagl et al. 1998). PvuI does not cut if the cytosines in its recognition sequence are methylated. Digestion of the plastid DNA with PvuI and hybridization to a 19.5 kb SalI fragment covering the ycf1-ndhB region of the tobacco plastid genome (Shinozaki et al. 1986) detects the expected four fragments of 24.5, 13, 5.5 and 2 kb in the wild-type (Nt-Wt). Note that the 5.5 kb fragment produces only a weak hybridization signal which is due to its limited coverage by the probe. Lack of any detectable DNA cleavage in the Nt-dcm plants suggests that the PvuI recognition sequences are efficiently methylated. M: DNA size marker (band sizes given in kb)