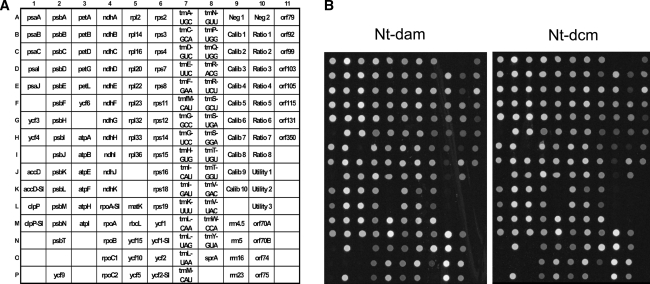
Fig. 3.
Analysis of expression of the plastid genome in Nt-dam and Nt-dcm transplastomic lines using the plastome microarray for Solanaceae (Kahlau and Bock 2008). a Spotting scheme of oligonucleotide probes on the microarray. The array contains all genes and conserved open reading frames (ycfs) present in Solanaceous plastid genomes and also a number of non-conserved open reading frames (orfs; for details see Kahlau and Bock 2008). Poorly conserved genes are covered by more than one oligonucleotide probe (indicated by the suffix ‘-S’ for Solanum-specific probe). NTC: negative control to assess non-specific background hybridization; Calib, Ratio, Utility: calibration and reference DNAs (artificial sequences corresponding to the Lucidea Universal ScoreCard; GE Healthcare; Kahlau and Bock, 2008). b Representative examples of microarray hybridization experiments for an Nt-dam and an Nt-dcm transplastomic line. No significant differences in expression of any plastid genome-encoded gene or open reading frame could be detected. Moreover, expression of the plastid genome in both Nt-dam and Nt-dcm was identical to the wild-type (not shown), indicating that neither adenine nor cytosine methylation affect plastid gene expression