Abstract
RNA enzymes have been developed that undergo self-sustained replication at a constant temperature in the absence of proteins1. These RNA molecules amplify exponentially through a cross-replicative process, whereby two enzymes catalyze each other’s synthesis by joining component oligonucleotides. Other RNA enzymes have been made to operate in a ligand-dependent manner by combining a catalytic domain with a ligand-binding domain (aptamer) to provide an “aptazyme”2,3. The principle of ligand-dependent RNA catalysis now has been extended to the cross-replicating RNA enzymes so that exponential amplification occurs in the presence, but not the absence, of the cognate ligand. The exponential growth rate of the RNA depends on the concentration of the ligand, enabling one to determine the concentration of ligand in a sample. This process is analogous to quantitative PCR (qPCR), but can be generalized to a wide variety of targets, including proteins and small molecules that are relevant to medical diagnostics and environmental monitoring.
A well-studied class of RNA enzymes are the RNA ligases, which catalyze the RNA-templated joining of RNA molecules. Some RNA ligases have been made to operate as aptazymes, and some of these have been made to undergo ligand-dependent catalytic turnover to provide linear signal amplification with ongoing target recognition4,5. One of the RNA ligases is the “R3C” RNA enzyme, which was obtained using in vitro evolution6. This enzyme has been reconfigured so that it can self-replicate by joining two RNA molecules that result in formation of another copy of itself7. It also has been converted to a cross-catalytic format, whereby two RNA enzymes catalyze each other’s synthesis from a total of four RNA substrates8. The cross-replication process is analogous to the ligase chain reaction9, except that in cross-replication the nucleic acid being amplified is itself the ligase, and strand separation occurs spontaneously without requiring temperature cycling.
The original cross-replicating RNA enzymes were slow catalysts that amplified poorly8. Recently their activity was substantially improved so that they can undergo efficient exponential amplification, generating about a billion copies in 30 h at a constant temperature of 42 °C1. Exponential amplification can be continued indefinitely, so long as a supply of the four substrates is maintained. The reaction requires 5–25 mM Mg2+, but does not require any proteins or other biological materials.
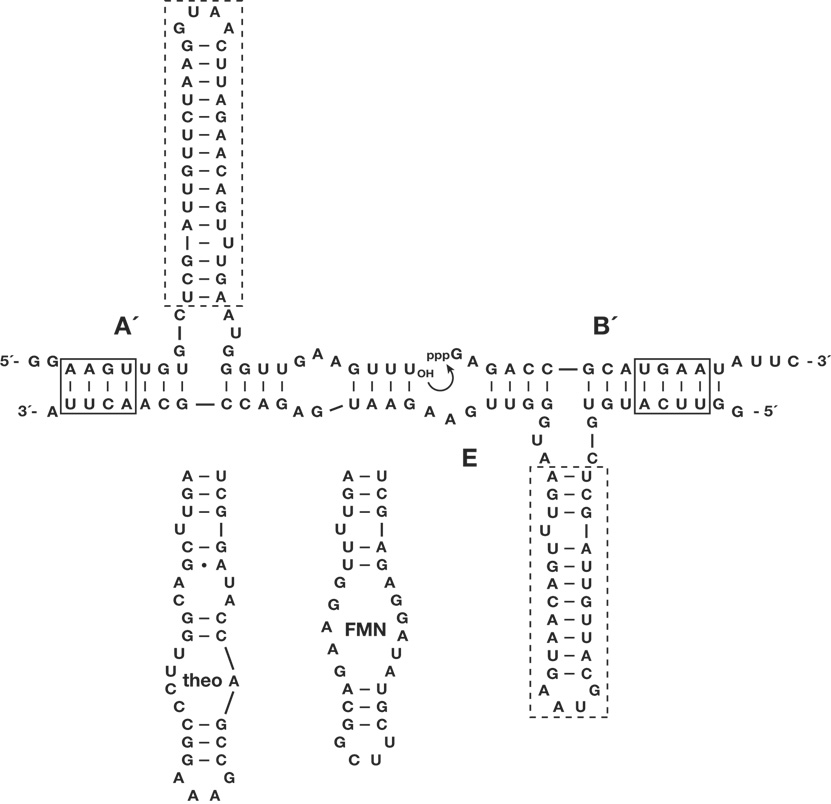
Cross-replication involves a plus-strand RNA enzyme (E) that catalyzes the joining of two substrates (A´ and B´) to form a minus-strand enzyme (E´), which in turn catalyzes the joining of two substrates (A and B) to form a new plus-strand enzyme (E). The cross-replicating enzymes were converted to aptazymes by replacing the distal portion of the central stem-loop by an aptamer that binds a particular ligand (Fig. 1). The aptamer was installed in the substrates A and A´, and in the corresponding enzymes E and E´. Two different aptamers were chosen, one that binds theophylline (theo)10 and another that binds flavin mononucleotide (FMN)11. In the absence of the ligand the aptamer domain is unstructured, resulting in destabilization of the adjacent catalytic domain, while in the presence of the ligand the catalytic domain becomes ordered so that exponential amplification can occur. The stability of the stem region connecting the aptamer and catalytic domains was adjusted to maximize the ratio of activity in the “on” (ligand present) compared to “off” (ligand absent) states. Unlike for conventional aptazymes, ligand-dependent activity is expressed exponentially in the growth rate of autocatalytic aptazymes, establishing sharp thresholds for ligand-dependent behavior.
Figure 1.
Sequence and secondary structure of autocatalytic aptazymes. The complex shown is that of the enzyme E and its substrates A´ and B´. Curved arrow indicates the site of ligation, resulting in formation of E´. The reciprocal reaction, involving the enzyme E´ and substrates A and B, is not shown. Dashed boxes indicate regions that were replaced by either the theophylline or FMN aptamer to form the corresponding aptazymes. Solid boxes indicate regions of Watson-Crick pairing that were replaced to allow multiplexed exponential amplification (the AAGU sequence in A´ was replaced by AGUA; the UGAA sequence in B´ was replaced by AUGA).
The two theophylline-dependent aptazymes, Etheo and E´theo, first were tested individually in a ligation reaction carried out under saturating conditions in the presence of 5 mM theophylline, exhibiting reaction rate constants of 1.4 and 0.6 min−1, respectively (Supplementary Fig. 1 online). Both enzymes had no detectable activity (<10−4 min−1) in the absence of theophylline or in the presence of 5 mM caffeine (which differs from theophylline by the presence of a methyl group at the N7 position of caffeine).
Cross-replication was initiated by adding 0.02 µM each of Etheo and E´theo to a reaction mixture containing 5 µM each of Atheo, A´theo, B, and B´, and either 5 mM theophylline or 5 mM caffeine, which was maintained at a constant temperature of 42 °C. Brisk exponential amplification occurred in the presence of theophylline, but there was no detectable amplification in the presence of caffeine (Fig. 2a). Exponential amplification resulted in the formation of new copies of both Etheo and E´theo, ultimately limited by the supply of substrates. A plot of enzyme concentration versus time exhibited a classic sigmoidal shape, indicative of exponential growth subject to a fixed supply of materials. These data were fit to the logistic growth equation:
where [E]t is the concentration of E (or E´) at time t, a is the maximum extent of growth, b is the degree of sigmoidicity, and c is the exponential growth rate.
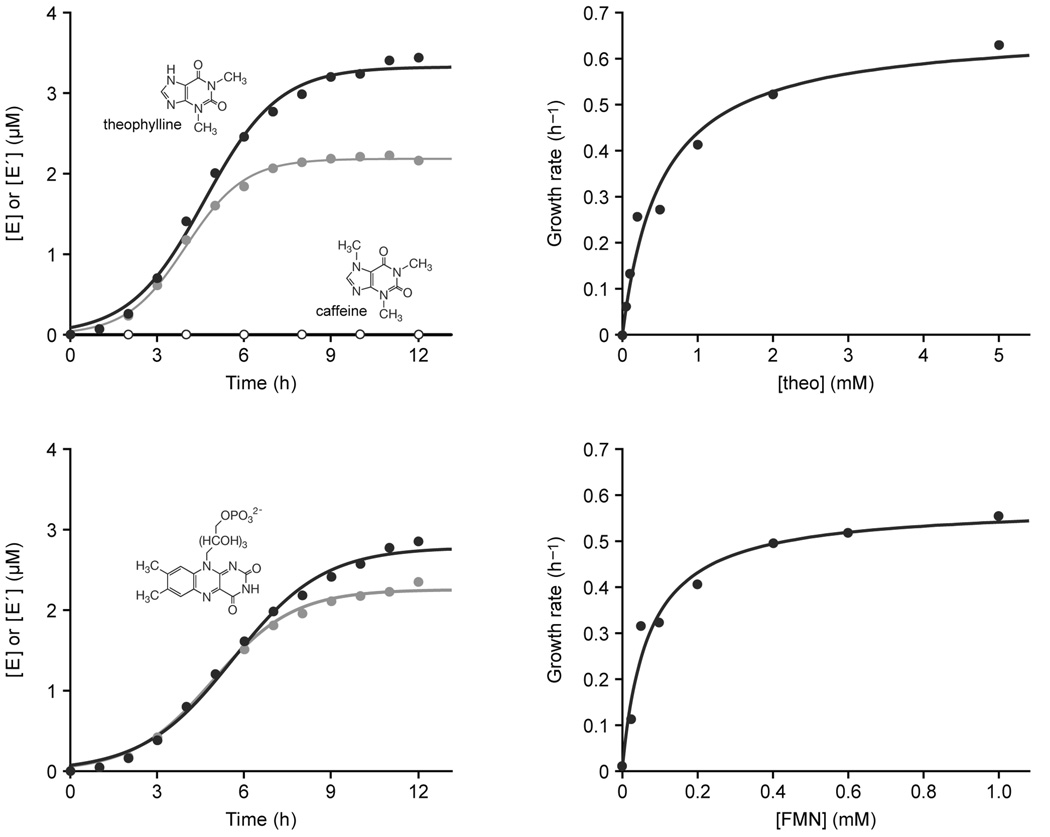
Figure 2.
Ligand-dependent exponential amplification of RNA. (a) The theophylline-dependent aptazymes, Etheo (black) and E´theo (gray), amplified exponentially in the presence of 5 mM theophylline (filled circles), but not in the presence of 5 mM caffeine (open circles). The structures of theophylline and caffeine are shown. (b) Exponential growth rate of Etheo in the presence of various concentrations of theophylline. (c) The FMN-dependent aptazymes, EFMN (black) and E´FMN (gray), amplified exponentially in the presence of 1 mM FMN. The structure of FMN is shown. (d) Exponential growth rate of EFMN in the presence of various concentrations of FMN. Growth rates for reactions that did not proceed beyond 10% fraction reacted were determined by a linear rather than exponential fit.
The exponential growth rates of Etheo and E´theo were 0.78 and 0.97 h−1, respectively, corresponding to a doubling time of about 50 min.
The maximum extents of synthesis of Etheo and E´theo were 3.3 and 2.2 µM, respectively. Exponential growth can be continued indefinitely, however, if a portion of the completed reaction mixture is transferred to a new mixture that contains a fresh supply of substrates. This is analogous to reseeding the PCR, but unlike the PCR remains dependent on the presence of the ligand throughout the amplification process, thus avoiding target-independent amplification. Following ~100-fold amplification, 1% of the reaction mixture was transferred to a new reaction vessel that contained 5 µM each of the four substrates, but only those enzymes that were carried over in the transfer. Three successive incubations were carried out in this manner, resulting in 106-fold overall amplification after 15 h (Supplementary Fig. 2 online).
The exponential growth rate of cross-replicating aptazymes is dependent on the concentration of the corresponding ligand. This allows one to construct standardized curves that can be used to determine the concentration of ligand in an unknown sample. The theophylline-dependent aptazymes were exposed to theophylline levels ranging from 0.2 to 5.0 mM and the exponential growth rate of Etheo was determined. The growth rate as a function of theophylline concentration provided a saturation curve (Fig. 2b), which revealed that the aptazyme binds theophylline with a Kd of 0.51 mM. Thus, the aptazyme can be used to measure theophylline concentrations in the range of approximately 0.05–5 mM. The Kd for the theophylline aptamer in isolation is 0.1 µM10, indicating that the aptamer is significantly destabilized in the context of the aptazyme. No attempt was made to optimize the aptamer in this context, as has been done for other aptazymes using in vitro selection12,13.
The FMN-dependent aptazymes also underwent exponential amplification in the presence, but not the absence, of their cognate ligand. The exponential growth rates of EFMN and E´FMN in the presence of 1 mM FMN were 0.58 and 0.70 h−1, respectively (Fig. 2c). The exponential growth rate of EFMN was determined in the presence of various concentrations of FMN, which provided a saturation curve (Fig. 2d) and revealed that the aptazyme binds FMN with a Kd of 0.068 mM. The same FMN aptamer has been linked to the hammerhead ribozyme and exhibited a Kd of 5 µM in that context12. This compares with a Kd of 0.5 µM for the FMN aptamer in isolation11.
Ligand-dependent exponential amplification can be performed using a pair of cross-replicating aptazymes that recognize two different ligands. As an example, a reaction was carried out employing 0.02 µM each of Etheo and E´FMN, and 5 µM each of Atheo, A´FMN, B, and B´. There was no amplification in the absence of both ligands, and only linear amplification in the presence of either theophylline or FMN, but robust exponential amplification in the presence of both ligands (Supplementary Fig. 3 online). This system can be regarded as performing a logical AND operation, providing exponential signal amplification that is dependent on the presence of two different inputs.
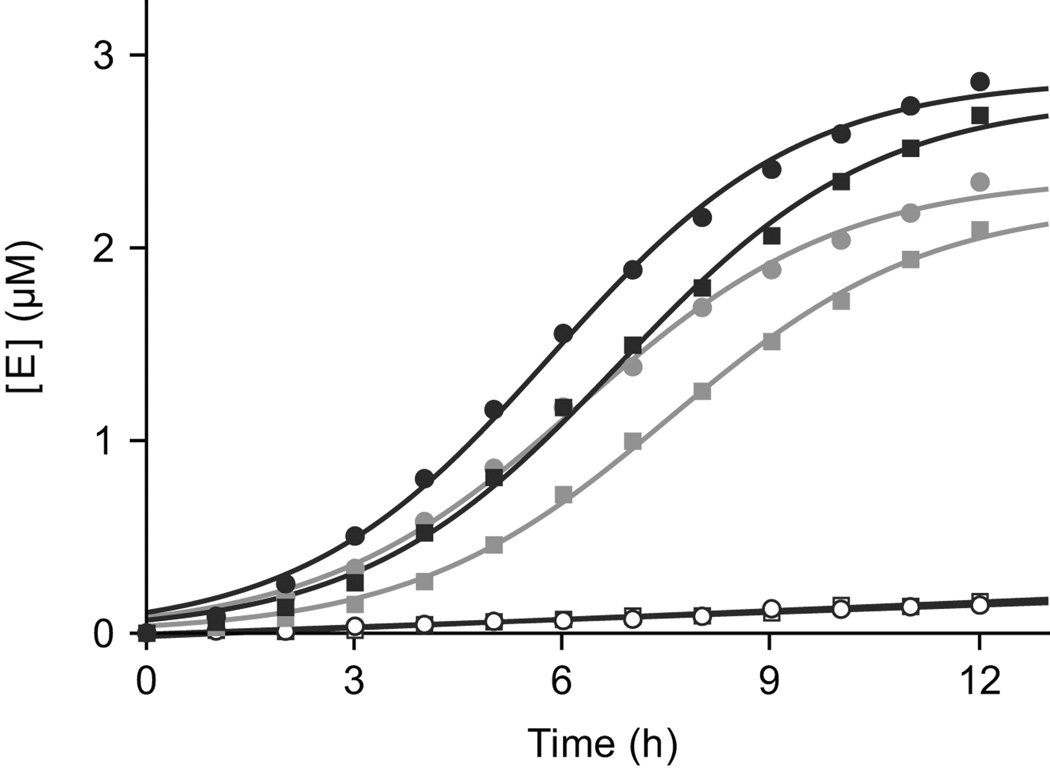
It is straightforward to carry out multiplexed ligand-dependent exponential amplification, employing two or more pairs of cross-replicating RNA enzymes that recognize their partners through distinct Watson-Crick pairing interactions (Fig. 1). Twelve pairs of cross-replicating RNA enzymes have been described1, one of which was chosen to contain the theophylline aptamer and another to contain the FMN aptamer (installing the same aptamer in both members of a cross-replicating pair). In the presence of either 5 mM theophylline or 0.7 mM FMN, only the corresponding RNA enzymes amplified exponentially, with growth rates for Etheo or EFMN of 0.35 or 0.43 h−1, respectively (Fig. 3). In the presence of both ligands, both pairs of cross-replicating enzymes amplified exponentially, with growth rates for Etheo and EFMN of 0.45 and 0.43 h−1, respectively.
Figure 3.
Multiplexed ligand-dependent exponential amplification of RNA. The theophylline-and FMN-dependent aptazymes were made to contain distinct regions of Watson-Crick pairing (Fig. 1). Exponential amplification of Etheo (circles) and EFMN (squares) occurred in the presence of both ligands (black) and in the presence of their cognate ligand alone (gray), but not in the presence of the non-cognate ligand alone (open symbols). Reaction mixtures contained 0.1 µM Etheo and E´theo, 0.02 µM EFMN and E´FMN, and 5 µM each of the eight corresponding RNA substrates.
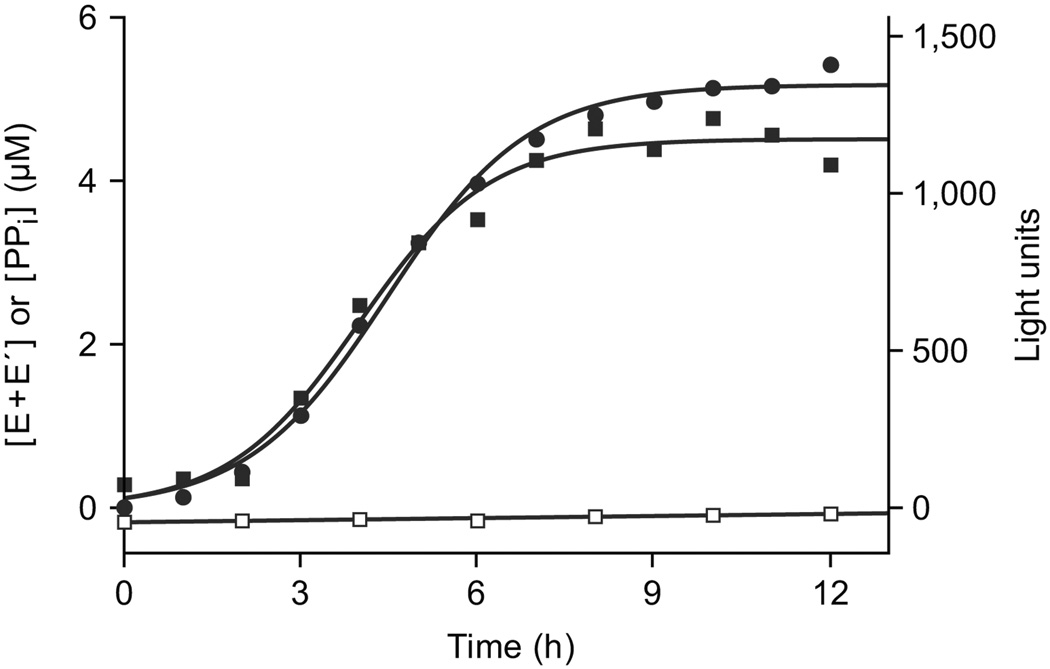
With each RNA-catalyzed ligation event, a 3´,5´-phosphodiester linkage is formed and one molecule of inorganic pyrophosphate is released6. The released pyrophosphate can be used to generate a luminescent signal based on an ATP-regenerative luciferase assay14. A plot of light emission over the course of theophylline-dependent exponential amplification was nearly identical to that for formation of the ligated products (Fig. 4). The luminescent signal generated by various known concentrations of pyrophosphate was used to determine a conversion factor for relating light units to absolute concentrations of pyrophosphate (Supplementary Fig. 4 online). These absolute concentrations were in close agreement with the absolute yield of ligated products over the course of exponential amplification (Fig. 4).
Figure 4.
Monitoring the course of exponential amplification by a luciferase assay, driven by the release of inorganic pyrophosphate that accompanies RNA ligation. Amplification was carried out in the presence of 5 mM theophylline, and the summed yields of Etheo and E´theo were measured both by separating the ligated products in a denaturing polyacrylamide gel (filled circles) and based on the luminescent signal generated by an ATP-regenerative luciferase assay14 (filled squares). Light units were converted to absolute concentrations of inorganic pyrophosphate based on comparison to known standards (Supplementary Fig. 4 online). There was no light signal above background in the absence of theophylline (open squares); slightly negative values are due to imprecision in determining the conversion factor.
The major limitation of autocatalytic aptazymes as a quantitative method for ligand-dependent exponential amplification is the need for the aptamer domain to bind its ligand with some requisite affinity, while remaining compatible with efficient cross-replication. The desired binding affinity usually is dictated by the concentration of the ligand in its biological or environmental context. Methods are well established for generating RNA aptamers that bind a target protein or small molecule with a particular affinity, often in the nanomolar range15,16. When these aptamers are placed in the context of an aptazyme, further optimization may be needed to regain the desired affinity. However, if the ligand concentration is very low, the concentration of RNA substrates required for efficient exponential amplification (typically micromolar) will exceed the desired Kd for the aptamer-ligand interaction. This would still allow ligand-dependent amplification, but at a reduced rate that is no longer dependent on the ligand concentration. One remedy would be to improve the Km of the cross-replicating enzymes so that the enzyme-substrate interactions remain saturated even when the aptamer-ligand interaction is unsaturated. However, this approach would limit the amount of signal that could be generated for very low-abundance targets. Another approach, analogous to qPCR and other methods that link a rare recognition event to subsequent exponential amplification17,18, would be to employ RNA replication as a reporter that is triggered by a recognition event. Unlike qPCR such a process would be isothermal, but like qPCR it would not benefit from ongoing sensing of the ligand during the course of exponential amplification.
Another limitation of autocatalytic aptazymes is that the molecules are composed of RNA, which is susceptible to degradation by ribonucleases or inhibition by non-specific RNA-binding proteins. The theophylline-dependent aptazymes were rapidly degraded in the presence of 10% bovine calf serum, but were able to undergo unimpeded ligand-dependent exponential amplification in the presence of serum that had been deproteinized by phenol extraction (Supplementary Fig. 5 online). Clearly it will be necessary to develop nuclease-resistant forms of the aptazymes, as has been done for most aptamers that are employed in a biological context19,20.
Aptamers and aptazymes have emerged as powerful tools for detecting and generating biochemical responses to a wide variety of ligands3. Nature has exploited this mechanism in the operation of “riboswitches”21, which are ligand-dependent riboregulators that occur widely in biology22. Scientists have engineered aptamers and aptazymes to sense proteins or small molecules23–26, to control gene expression27–28, and to perform molecular computation29. Autocatalytic aptazymes may be useful in some of these applications because they provide both specificity through dynamic sensing of the ligand and sensitivity due to ligand-dependent exponential amplification. Although several practical concerns still must be addressed, the ability to perform quantitative analysis of a variety of ligands under isothermal conditions may have utility in medical diagnostics and environmental monitoring.
Methods
Materials
Oligonucleotides were synthesized on an Expedite automated DNA/RNA synthesizer (Applied Biosystems, Foster City, CA) using nucleoside phosphoramidites purchased from Glen Research (Sterling, VA). All oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and desalted using a C18 SEP-Pak cartridge (Waters, Milford, MA). Histidine-tagged T7 RNA polymerase was purified from E. coli strain BL21 containing plasmid pBH161 (kindly provided by William McAllister, State University of New York, Brooklyn). Thermus aquaticus DNA polymerase was cloned from total genomic DNA and purified as described previously30. M1 RNA, the catalytic subunit of RNAse P, was obtained from E. coli genomic DNA (Sigma-Aldrich, St. Louis, MO) by PCR amplification and subsequent in vitro transcription, as described previously1. Calf intestine phosphatase and T4 polynucleotide kinase were purchased from New England Biolabs (Ipswich, MA), yeast inorganic pyrophosphatase was from Sigma-Aldrich, and bovine pancreatic DNase I was from Roche Applied Science (Indianapolis, IN). Nucleoside and deoxynucleoside 5´-triphosphates, theophylline, and FMN were purchased from Sigma-Aldrich, [γ-32P]ATP (7 µCi/pmol) was from Perkin Elmer (Waltham, MA), and caffeine was from MP Biomedicals (Solon, OH). Photinus pyralis (firefly) luciferase, Saccharomyces cerevisiae adenosine-5´-triphosphate sulfurylase, adenosine 5´-phosphosulfate, and d-luciferin were from Sigma-Aldrich. Bovine calf serum was from Omega Scientific (Tarzana, CA) and Superasin (RNAse inhibitor) was from Ambion (Austin, TX).
Preparation of aptazymes and substrates
All RNA enzymes and substrates were prepared by in vitro transcription in a reaction mixture containing 0.4 µM DNA template, 0.8 µM synthetic oligodeoxynucleotide having the sequence 5´-GGACTAATACGACTCACTATA-3´ (T7 RNA polymerase promoter sequence underlined), 2 mM each of the four NTPs, 15 U/µL T7 RNA polymerase, 0.001 U/µL inorganic pyrophosphatase, 15 mM MgCl2, 2 mM spermidine, 5 mM dithiothreitol, and 50 mM Tris-HCl (pH 7.5). The mixture was incubated at 37 °C for 2 h, quenched by adding an equal volume of 15 mM Na2EDTA, treated with 1 U/µL DNase I, and extracted with a 1:1 mixture of phenol:chloroform. The RNA was precipitated, purified by PAGE, and desalted. Transcription of M1 RNA was performed similarly, except employing a double-stranded DNA template that was generated by PCR.
The A and A´ substrates could not be obtained reliably by in vitro transcription due to heterogeneity at the 3´ end of the transcripts. Instead, these substrates were prepared from the corresponding E or E´ molecules by cleaving off the B or B´ portion using E. coli M1 RNA, as described previously1. The external guide sequence RNA for cleavage of Etheo and EFMN had the sequence 5´-CGUAAGUUGCGGUCUCACCA-3´, and for E´theo and E´FMN had the sequence 5´-AUAUUCAUGCGGUCUCACCA-3´ (nucleotides complementary to the target RNA underlined). For the second pair of Etheo and E´theo molecules used in the multiplex experiments, the external guide sequence RNAs had the sequence 5´-CGUAGUAUGCGGUCUACCA-3´ and 5´-GAAUAUCAUUGCGGUCUCACCA-3´, respectively. The A and A´ substrates were [5´-32P]-labeled by first dephosphorylating using calf intestine alkaline phosphatase, then phosphorylating using T4 polynucleotide kinase and [γ-32P]ATP. The labeled substrates were purified by PAGE and desalted using a Nensorb 20 cartridge (NEN Life Sciences, Waltham, MA).
Individual RNA-catalyzed reactions
RNA-catalyzed RNA ligation was performed in a reaction mixture containing 5 µM E or E´, 0.1 µM [5´-32P]-labeled A´ or A, 6 µM B´ or B, 25 mM MgCl2, and 50 mM EPPS (pH 8.5), which was incubated at 42 °C. Aliquots were taken at various times and quenched by adding an equal volume of gel-loading buffer containing 50 mM Na2EDTA and 18 M urea. The products were separated by PAGE and quantitated using a PharosFX molecular imager (Bio-Rad, Hercules, CA). The data were fit to the equation:
where Ft is the fraction reacted at time t, Fmax is the overall maximum extent of the reaction, a1 and k1 are the amplitude and rate of the initial fast phase, and a2 and k2 are the amplitude and rate of the subsequent slow phase, respectively.
In the presence of 5 mM theophylline, the reaction catalyzed by Etheo exhibited a fast phase with an amplitude of 0.57 and rate constant of 1.4 min−1, followed by a slow phase with an amplitude of 0.24 and rate constant of 0.044 min−1; the reaction catalyzed by E´theo had an amplitude of 0.52 and rate constant of 0.59 min−1 in the fast phase, and an amplitude of 0.26 and rate constant of 0.045 min−1 in the slow phase.
Cross-replication reactions
Cross-catalytic exponential amplification was performed in a reaction mixture containing 0.02 µM each of E and E´, 5 µM each of [5´-32P]-labeled A and A´, 5 µM each of B and B´, 25 mM MgCl2, and 50 mM EPPS (pH 8.5), which was incubated at 42 °C. The reaction was initiated by mixing equal volumes of two solutions, one containing the enzymes and substrates, and the other containing the MgCl2 and EPPS buffer. Aliquots were taken at various times, quenched, and the amounts of newly-synthesized E and E´ were quantitated as described above. The data were fit to the logistic growth equation, as described in the main text.
Luciferase assays
Known concentrations of inorganic pyrophosphate or samples taken from the cross-replication reaction were diluted 10-fold into a reaction mixture containing 0.15 µg/µL luciferase, 0.00045 U/µL ATP sulfurylase, 10 µM adenosine 5´-phosphosulfate, 0.5 mM d-luciferin, 25 mM magnesium acetate, 0.1% bovine serum albumin, 1 mM dithiothreitol, 0.4 µg/µL polyvinylpyrrolidone (MW 360,000), and 100 mM Tris-acetate (pH 7.75). The pyrophosphate standards were prepared in a solution identical to that employed in cross-replication, but lacking the RNA enzymes and substrates. Luminescence was detected using a Perkin Elmer LS55 luminescence spectrometer operating in bioluminescence mode, with a PMT voltage of 900 V, cycle time of 200 ms, gate time of 180 ms, and delay time of 0. The flash count was set to 1, the emission filter was fully open, and the emission slit width was 12 nm. Following addition of the sample to the luciferase mixture, luminescence was monitored for 5 min with a 0.1 s integration time. The amount of light generated was linear over a pyrophosphate concentration range of 0.1–10 µM.
Supplementary Material
Note: Supplementary information is available on the Nature Biotechnology website.
Acknowledgments
This work was supported by grant R01GM065130 from the NIH and by The Skaggs Institute for Chemical Biology at The Scripps Research Institute. B.J.L. was supported by NIH Ruth L. Kirschstein National Research Service Award 5F32GM078691.
Footnotes
Competing interests statement. A patent application has been filed describing the method of ligand-dependent exponential amplification of RNA.
References
- 1.Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009 Jan 8; doi: 10.1126/science.1167856. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem. Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 3.Hesselberth JR, Robertson MP, Knudsen SM, Ellington AD. Simultaneous detection of diverse analytes with an aptazyme ligase array. Anal. Biochem. 2003;312:106–112. doi: 10.1016/s0003-2697(02)00441-4. [DOI] [PubMed] [Google Scholar]
- 4.Hartig JS, et al. Protein-dependent ribozymes report molecular interactions in real time. Nat. Biotechnol. 2002;20:717–722. doi: 10.1038/nbt0702-717. [DOI] [PubMed] [Google Scholar]
- 5.Vaish NK, et al. Monitoring post-translational modification of proteins with allosteric ribozymes. Nat. Biotechnol. 2002;20:810–815. doi: 10.1038/nbt719. [DOI] [PubMed] [Google Scholar]
- 6.Rogers J, Joyce GF. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul N, Joyce GF. A self-replicating ligase ribozyme. Proc. Natl. Acad. Sci. USA. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D-E, Joyce GF. Cross-catalytic replication of an RNA ligase ribozyme. Chem. Biol. 2004;11:1505–1512. doi: 10.1016/j.chembiol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Wu DY, Wallace RB. The ligation amplification reaction (LAR) — amplification of specific DNA sequences using sequential rounds of template-dependent ligation. Genomics. 1989;4:560–569. doi: 10.1016/0888-7543(89)90280-2. [DOI] [PubMed] [Google Scholar]
- 10.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 11.Burgstaller P, Famulok M. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew. Chemie. 1994;33:1084–1087. [Google Scholar]
- 12.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc. Natl. Acad. Sci. USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson MP, Ellington AD. Design and optimization of effector-activated ribozyme ligases. Nucleic Acids Res. 2000;28:1751–1759. doi: 10.1093/nar/28.8.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 15.Fitzwater T, Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- 16.Ciesiolka J, et al. Affinity selection-amplification from randomized ribooligonucleotide pools. Methods Enzymol. 1996;267:315–335. doi: 10.1016/s0076-6879(96)67021-9. [DOI] [PubMed] [Google Scholar]
- 17.Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 18.Fredriksson S, et al. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Qiu Q, Gill SC, Jayasena SD. Modified RNA sequence pools in in vitro selection. Nucleic Acids Res. 1994;22:5229–5234. doi: 10.1093/nar/22.24.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green LS, et al. Nuclease-resistant nucleic acid ligands to vascular permeability factor/vascular endothelial growth factor. Chem. Biol. 1995;2:683–695. doi: 10.1016/1074-5521(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 21.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 22.Mandal M, Breaker RR. Gene regulation by riboswitches. Nature Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 23.Nutiu R, Li Y. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 24.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 25.Bock C, et al. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics. 2004;4:609–618. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- 26.Kirby R, et al. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal. Chem. 2004;76:4066–4075. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- 27.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule–RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 28.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida W, Yokobayashi Y. Photonic boolean logic gates based on DNA aptamers. Chem. Commun. 2007;2007:195–197. doi: 10.1039/b613201d. [DOI] [PubMed] [Google Scholar]
- 30.Pluthero FG. Rapid purification of high-activity Taq DNA polymerase. Nucleic Acids Res. 1993;21:4850–4851. doi: 10.1093/nar/21.20.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Biotechnology website.