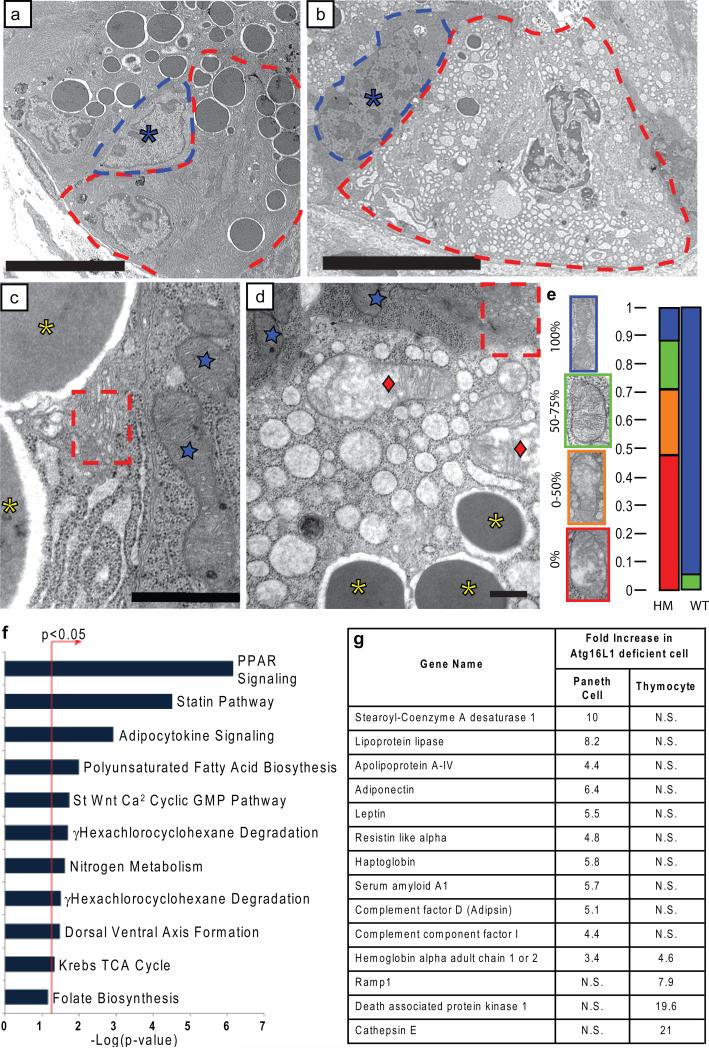
Figure 3. Critical role of Atg16L1 in the structure and transcriptional profile of Paneth cells.
a-d, EM of littermate WT (a, c) and Atg16L1HM (b, d) Paneth cells (a, b: dotted lines denote individual cells, asterisk indicates normal progenitor cell; c, d: blue stars indicate normal mitochondria in WT Paneth cells and Atg16L1HM progenitor cells, red diamonds indicate degenerating mitochondria, dotted Box shows normal Golgi compartments, and yellow asterisks indicate granules) (n = 3 mice/genotype). e, Mitochondrial integrity was quantified based on the percentage of the visible mitochondrial section displaying intact cisternae (n = 49 WT and 71 Atg16L1HM cells). f, Enrichment of pathways associated with human orthologs mapped from genes differentially expressed in Atg16L1 deficient Paneth cells relative to WT cells, using the canonical pathway collection from MSigDB24. The bar chart displays the negative log of the enrichment p-values for each pathway using the hypergeometric distribution (see methods). *denotes pathways found to be significantly enriched (p<0.05). Pathways with only one gene assigned were excluded from the chart. g, Chart of selected genes whose mRNA are enriched in either Atg16L1 deficient Paneth cells (baseline = WT Paneth cells) or Atg16L1 deficient thymocytes (baseline = WT thymocytes). N.S. = not significant. Scale bars: a-b, 10 μm; c-d, 500nm.