Abstract
Nested clade phylogeographic analysis (NCPA) and approximate Bayesian computation (ABC) have been used to test phylogeographic hypotheses. Multilocus NCPA tests null hypotheses, whereas ABC discriminates among a finite set of alternatives. The interpretive criteria of NCPA are explicit and allow complex models to be built from simple components. The interpretive criteria of ABC are ad hoc and require the specification of a complete phylogeographic model. The conclusions from ABC are often influenced by implicit assumptions arising from the many parameters needed to specify a complex model. These complex models confound many assumptions so that biological interpretations are difficult. Sampling error is accounted for in NCPA, but ABC ignores important sources of sampling error that creates pseudo-statistical power. NCPA generates the full sampling distribution of its statistics, but ABC only yields local probabilities, which in turn make it impossible to distinguish between a good fitting model, a non-informative model, and an over-determined model. Both NCPA and ABC use approximations, but convergences of the approximations used in NCPA are well defined whereas those in ABC are not. NCPA can analyze a large number of locations, but ABC cannot. Finally, the dimensionality of tested hypothesis is known in NCPA, but not for ABC. As a consequence, the “probabilities” generated by ABC are not true probabilities and are statistically non-interpretable. Accordingly, ABC should not be used for hypothesis testing, but simulation approaches are valuable when used in conjunction with NCPA or other methods that do not rely on highly parameterized models.
Keywords: nested clade analysis, phylogeography, statistics, Bayesian analysis, computer simulation analysis
Intraspecific phylogeography is the investigation of the evolutionary history of populations within a species over space and time. This field entered its modern era with the pioneering work of Avise and co-workers (Avise et al. 1987; 1979) who used haplotype trees as their primary analytical tool. A haplotype tree is the evolutionary tree of the various haplotypes found in a DNA region of little to no recombination. The early phylogeographic studies made qualitative inference from a visual overlay of the haplotype tree upon a map of the sampling locations. As the field matured, there was a general recognition that the inferences being made were subject to various sources of error, and the next phase in the development of intraspecific phylogeography was to integrate statistics into the inference structure.
One of the first statistical phylogeographic methods was nested clade phylogeographic analysis (NCPA) (Templeton et al. 1995). This method incorporates the error in estimating the haplotype tree (Templeton et al. 1992; Templeton& Sing 1993) and the sampling error due to the number of locations and the number of individuals sampled (Templeton et al. 1995). Since 2002, nested clade analysis has been radically modified and extended to take into account the randomness associated with the coalescent and mutational processes; to minimize false positives and type I errors through cross validation, and to test specific phylogeographic hypotheses through a likelihood ratio testing framework (Gifford& Larson 2008; Templeton 2002b, 2004a; Templeton 2004b). Statistical approaches to phylogeography have also been developed through simulations of specific phylogeographic models for both hypothesis testing and parameter estimation (Knowles 2004).
Beaumont and Panchal (2008) have questioned the validity of NCPA and held up the approximate Bayesian computation (ABC) method (Beaumont et al. 2002; Pritchard et al. 1999) as an alternative. They specifically cite the application of ABC by Fagundes et al. (2007) as an example of how tests of phylogeographic hypotheses should be performed. Therefore, I will compare the statistical properties of the ABC method to those of NCPA with a focus upon the work of Fagundes et al. (2007).
Strong versus Weak Inference
The two main types of statistical hypothesis testing are 1) testing a null hypothesis and 2) assessing the relative fit of alternative hypotheses. All inferences in NCPA start with the rejection of the null hypothesis of no association between haplotype clades with geography. When this null hypothesis is rejected, a biological interpretation of the rejection is sometimes possible through the application of an inference key (discussed in the next section). Knowles and Maddison (2002) pointed out that these interpretations from the inference key were not phrased as testable hypotheses. This deficiency of single-locus NCPA has been corrected in multilocus NCPA (Templeton, 2002; Templeton, 2004a; Templeton, 2004b). All inferences in multilocus NCPA are phrased as testable null hypotheses, and the only inferences retained are those that are confirmed by explicit statistical tests. The multilocus likelihood framework also allows many other specific phylogeographic models to be phrased as testable null hypotheses. For example, the out-of-Africa replacement hypothesis that human populations expanded out of Africa around 100,000 years ago and drove to complete genetic extinction all Eurasian human populations can be phrased as a testable null hypothesis within multilocus NCPA. This null hypothesis is decisively rejected with a probability less than 10−17 with data from 25 human gene regions using a likelihood ratio test (Templeton 2005, 2007b).
Nested clade analysis can also test null hypotheses about a variety of other data types. Indeed, nested clade analysis was first developed for testing the null hypothesis of no association between phenotypic variation with the haplotype variation at a candidate locus (Templeton et al. 1987). This robustness allows one to integrate hypothesis testing on morphological, behavioral, physiological, and ecological data with the phylogeographic analysis (e.g.,Templeton 2001; Templeton et al. 2000b), despite erroneous claims to the contrary (Lemmon& Lemmon 2008). Moreover, the same multilocus statistical framework can test null hypothesis about correlated phylogeographic events in different species, as illustrated by the likelihood ratio test that humans and their malarial parasite shared common range expansions (Templeton 2004a, 2007b). The criticism that single-locus interpretations are not testable hypotheses is true but increasingly irrelevant as the entire field of phylogeography moves towards multilocus data sets. Given that the Fagundes et al. (2007) is a multilocus analysis, the legitimate comparison is multilocus ABC versus multilocus NCPA and not with the pre-2002 single-locus version of NCPA criticized by Beaumont and Panchal (2008).
In contrast to multilocus NCPA, the ABC method posits two or more alternative hypotheses and tests their relative fits to some observed statistics. For example, Fagundes et al. (2007) used ABC to test the relative merits of the out-of-Africa replacement model of human evolution and two other models of human evolution (assimilation and multiregional). Of these three models of human evolution, the out-of-Africa replacement model had the highest relative posterior probability of 0.781.
Karl Popper (1959) argues that the scientific method cannot prove something to be true, but it can prove something to be false. In Popper's scheme, falsifying a hypothesis is strong scientific inference. The falsification of the replacement hypothesis by NCPA is an example of strong inference. Contrasting alternative hypotheses can also be strong inference when the alternatives exhaustively cover the hypothesis space. When the hypothesis space is not exhaustively covered, testing the relative merits among a set of hypotheses results in weak inference. A serious deficiency of weak inference occurs when all of the hypotheses being compared are false. It is still possible that one of them fits the data much better than the alternatives and as a result could have a high relative probability. The three basic models of human evolution considered by Fagundes et al. (2007) do not exhaustively cover the hypothesis space. For example, the model of human evolution that emerges from multilocus NCPA (Templeton 2005, 2007b) has elements from the out-of-Africa, assimilation, and multiregional models as well as completely new elements, such as an Acheulean out-of-Africa expansion around 650,000 years ago that is strongly corroborated by fossil, archeological and paleo-climatic data. Indeed, all the models given by Fagundes et al. (2007) have already been falsified by multilocus NCPA, so having a high relative probability does not mean that a hypothesis is true or supported by the data. The fact that the out-of-Africa replacement hypothesis is rejected with a probability of less than 10−17 is compatible with the out-of-Africa replacement hypothesis having a probability of 0.781 relative to two alternatives that have also been falsified. In the Popperian framework, the strong falsification of the out-of-Africa replacement has precedent over the weak relative support for the out-of-Africa replacement model against other falsified alternatives.
It is important to note that the difference between strong and weak inference is separate from the difference between likelihood and Bayesian statistics. Both likelihood and Bayesian methods can and have been used for both strong and weak inference. Indeed, NCPA uses both Bayesian and likelihood approaches. The haplotype network that defines the nested design is estimated (with explicit error) by the procedure now known as statistical parsimony, which quantifies the probability of deviations from parsimony by a Bayesian procedure (Templeton et al. 1992).
In general, there are always many possible combinations of fragmentation events, gene flow processes, expansions, and their times so as to preclude an exhaustive set of alternative hypotheses. Accordingly, the first distinction between multilocus NCPA and ABC is that multilocus NCPA can yield strong inference whereas ABC yields weak inference. Weak inference also means that ABC can give strong relative support to a hypothesis that is false, and there is no way within ABC of identifying or correcting for this source of false positives.
Interpretative Criteria
A statistical test returns a probability value, but rarely is the probability value per se the reason for an investigator performing the test. In such circumstances, statistical tests need to be interpreted. Such interpretation is part of the inference process for both NCPA and ABC.
NCPA provides an explicit interpretative key that is based upon predictions from neutral coalescent theory coupled with considerations from the sampling design. Because of explicit, a priori criteria, biological interpretations in NCPA are based on the same criteria for all species. The explicit nature of the interpretations has also allowed many users to comment upon ways of improving the interpretation of the statistical results and to allow the interpretations to be extensively validated by a set of 150 positive controls (Templeton 2004b, 2008). As a result, the interpretative key is dynamic and open-sourced, constantly being improved and subject to re-evaluation, just as the ABC method has been revised over the years. The interpretative key focuses upon the simple types of processes and events that operate in evolutionary history. A statistical analysis of the 150 positive controls also showed that there is no statistical interference among the components (Templeton 2004b). The overall phylogeography of the sampled organism emerges as these simple individual processes and events are put together. As a consequence, NCPA allows the discovery of unexpected events and processes and the inference of complex scenarios from simple events.
The use of positive controls to validate the inference key also revealed that the false positive rate of single-locus NCPA exceeded the nominal 5% level (Templeton 1998; Templeton 2004b). One of the prime motivations for the development of multilocus NCPA was to eliminate the false positives from single-locus inferences through another series of statistical tests that cross-validate the initial set of single-locus inferences. The false positive rates for single locus NCPA are therefore irrelevant to multilocus NCPA. These additional tests also mean that every inference in multilocus NCPA has been treated as a testable null hypothesis within a maximum likelihood framework.
The biological interpretations in ABC are defined by the finite set of models that are simulated: no biological interpretations outside of this set are allowed. This interpretative set is defined in an ad hoc, case-by-case basis. The interpretative set in ABC consists of models that specify an entire phylogeographic history rather than just phylogeographic components, as in NCPA. Because the interpretations are strictly limited to a handful of fully pre-specified phylogeographic models, no novelty or discovery is allowed in ABC.
An illustration of the sensitivity of the simulation approaches to the interpretative set is provided by a contrast of Ray et al. (2005) versus Eswaran et al. (2005). Both papers used computer simulations to measure the goodness of fit of several models of human evolution, including the out-of-Africa replacement model. However, Eswaran et al. (2005) included in their interpretative set a model of isolation by distance with a small number of genes under natural selection, a model not included in the interpretive set of Ray et al. (2005). Whereas Ray et al. (2005) concluded that the out-of-Africa replacement model was by far the best fitting model, Eswaran et al. (2005) concluded that the isolation-by-distance/selection model was so superior that the replacement model was refuted. Indeed, Eswaran et al. (2005) estimated that 80% of the loci in the human genome were influenced by admixture versus the 0% of the replacement model. Such dramatically different conclusions are not incompatible with one another because the interpretative sets were different in these two studies. This heterogeneity in relative fit as a function of different interpretative sets is an unavoidable feature of weak inference.
Another disadvantage of ABC is that the interpretative set cannot be validated, unlike the interpretative key of NCPA. First, positive controls cannot be used to validate ABC. The interpretations in NCPA are highly amenable to validation via positive controls because the units of inference are individual processes or events and not the entire phylogeographic history of the species under study. For example, suppose a species today occupies in part a region that was under a glacier 20,000 years ago. In such a case, one can be confident that the range of the species expanded into this formerly glaciated region sometime in the last 20,000 years. Consequently, this species can be used as a positive control for the inference of range expansion. There may be many other aspects of the species’ phylogeographic history, but a complete knowledge of the species phylogeography is not needed for it to be used as a positive control in NCPA. In contrast, ABC specifies the entire phylogeographic history of the sample under consideration. Although prior knowledge of specific events is commonplace, prior knowledge of the complete phylogeography of a species is rare, making ABC less amenable to positive controls than NCPA. There is also a logical difficulty to using positive controls with ABC. The only way for ABC to yield the correct phylogeographic model is to have the correct phylogeographic model in the interpretive set. Since by definition one knows the correct model in a positive control, this is easy to achieve, but it circumvents the primary cause of false positives in ABC: failing to include the correct model in the interpretative set. This same logical problem also means that computer simulations cannot determine the false positive rate of ABC. Because NCPA does not pre-specify its inference, false positive rates are easily determined (Templeton 1998; Templeton 2004b) and therefore corrections can and have been implemented: modifications of the original inference key, the development of multiple test corrections for single locus NCPA for both inferences across nesting clades and (if desired) within nesting clades (Templeton 2008), and the development of multilocus NCPA that eliminates false positives through cross-validation. This ability to quantify and deal with false positives is a large statistical advantage of NCPA over any method of weak inference in which the false positive rate cannot be determined even in principle.
The Impact of Implicit Assumptions
Another advantage of NCPA is its transparency. Single-locus NCPA is based on testing simple null hypotheses using well defined and long established permutational procedures (Edgington 1986). The inference key is explicit. Multilocus NCPA takes the inferences emerging from multiple single locus studies on the same species and subjects them to an explicit cross-validation and hypothesis testing procedure. The multilocus tests are based on explicit probability distributions and likelihood ratios, a standard and well-established statistical procedure. The complexity of the final multilocus NCPA model, such as that for human evolution (Templeton 2005; Templeton 2007a; Templeton 2007b), is built up from simple inferences, with each individual inference being tested as a null hypothesis in the cross validation procedure, making it obvious how the final model was achieved and its statistical support.
In contrast, the interpretive set for ABC and other simulation approaches start with the final, complex phylogeographic models that by necessity contain many assumptions and parameters in a confounded manner. For example, Fagundes et al. (2007) estimated the 95% highest posterior density (a Bayesian analogue of a 95% confidence interval) of the admixture parameter between archaic Africans with archaic Eurasians to be 6.3 × 10−5 to 0.023 under their assimilation model. This statistical claim is based on only 8 east Asian individuals to represent all of Eurasia (Eurasia is the potentially admixed population) and 50 loci. Such small admixture values are difficult to estimate accurately, and current admixture studies generally recommend samples in the several hundreds with thousands of loci (Bercovici et al. 2008). The sample sizes are so small and the geographical sampling so sparse that the data set given by Fagundes et al. (2007) does not achieve the minimums given in Templeton (2002b) for testing the out-of-Africa hypothesis with NCPA. I was only able to replicate their claims about the admixture parameter if I assumed that archaic Eurasians and archaic Africans were highly genetically differentiated and treated the eight East Asians as the population of inference rather than being a sample from the Eurasian human population (more on sampling in the next section).
This assumption of extreme genetic differentiation between archaic Africans and Asians was confirmed by one of the coauthors (Excoffier, personal communication), who explained that it arose from information in Figure 1 in the published paper coupled with information about parameter values in Table 7 from the online supplementary material. Figure 1 in Fagundes et al. (2007) shows that they modeled archaic Africans and archaic Eurasians as being completely isolated populations for a long period of time before the admixture event. Supplementary Table 7 shows that they assumed that this period of isolation began between 32,000 to 40,000 generations ago from the present and lasted until the out-of-Africa expansion that they modeled as occurring between 1,600 to 4,000 generations ago. Assuming a generation time of 20 years for these archaic populations (Takahata et al. 2001), this translates into a period of genetic isolation lasting between 560,000 to 768,000 years during which genetic differences could accumulate. Extreme genetic differentiation is ensured by their assumptions of small population size (an average of 5,050) in both archaic populations. These assumed population sizes would result in an average coalescence time for an autosomal locus (4N) of 20,100 generations (402,000 years). This average coalescence time is shorter than the interval of isolation, which is essential to create high levels of genetic divergence. Moreover, they assumed that the initial colonization of Eurasia by Homo erectus involved between 2 to 5,000 individuals, and this bottleneck would further reduce coalescence times in the archaic Eurasian population and enhance genetic differentiation between archaic Eurasians and archaic Africans. These choices of parameter values produce the extreme genetic differentiation that is necessary to obtain their published confidence interval on the admixture rate from only 8 individuals and 50 loci. However, these same parameter values lead to discrepancies with what is known about coalescence times of autosomal loci in humans. For example, Figure 3 in Templeton (2007a) shows the estimated coalescence times of eleven autosomal loci, all of which are greater than 402,000 years, and indeed 10 of the 11 have coalescence times greater than one million years. Similarly, Takahata et al. (2001) estimated the coalescence times of four autosomal loci, all of which were greater than 402,000 years, and two of which were greater than a million years. Takahata et al. (2001) used a 5 MYA (million years ago) calibration date for the human/chimpanzee split, whereas this split is now commonly put at 6 MYA because of better fossil data. Using the newer calibration point, 3 out the 4 autosomal loci in Takahata's analysis now have coalescence times greater than 1 MYA. In either case, it is patent that the parameter values chosen by Fagundes et al. (2007) are strongly discrepant with the empirical data on autosomal coalescence times.
Figure 1.
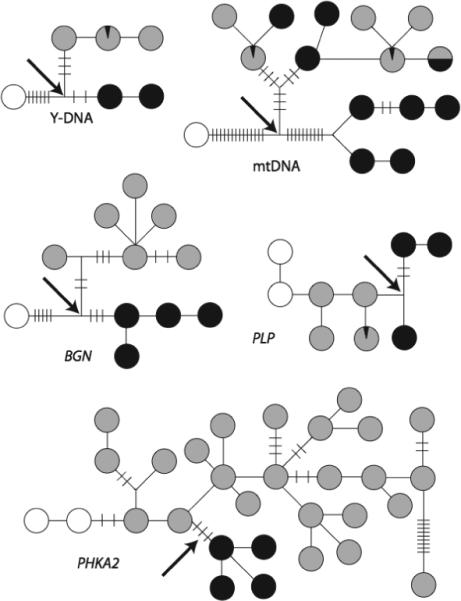
Five haplotype trees estimated from samples of African savanna elephants (black), African forest elephants (grey), and Asian elephants as an outgroup (white circles) by Roca et al. (2005). A black arrow shows the points in the haplotype trees at which a significant fragmentation event was inferred by NCPA that primarily separates the forested and savanna areas of Africa.
Figure 3.
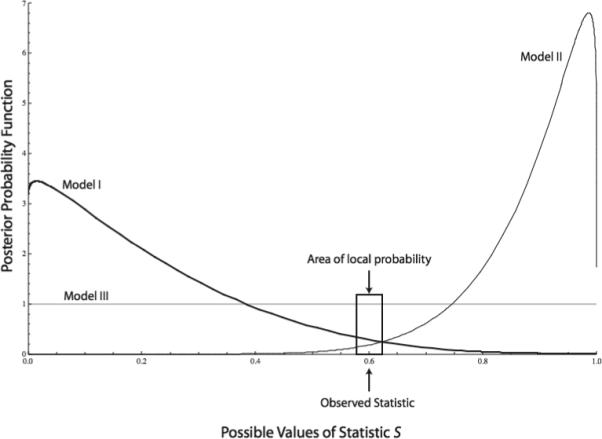
Hypothetical posterior distributions for three models for a univariate statistic and an area around the observed value of statistic where local probabilities are evaluated.
There are two basic reasons for rejecting a model in ABC. One, the model is wrong (or at least more wrong than the alternatives to which it is compared); and two, the biological model is correct but the simulated parameter values are wrong. It is not clear if the rejection of the assimilation model by Fagundes et al. (2007) is due to the assimilation model being wrong or is due to the unrealistic parameter values they chose for this model. These two causes of poor fit are confounded in ABC, thereby preventing meaningful biological interpretations of the rejection.
The rejection of the assimilation model by Fagundes et al. (2007) reveals another fundamental weakness of the ABC inference structure. Although Fagundes et al. (2007) interpreted their rejection of the assimilation model in terms of its admixture component, their assimilation model also includes the long period of prior isolation between archaic Eurasians and Africans. There is no necessity to couple admixture with a long period of complete genetic isolation. Indeed, in the model of human evolution that emerges from multilocus NCPA, there is both admixture and gene flow with no extended period of isolation. ABC simulates the entire, complex phylogeographic scenario as a single entity. Thus, the rejection of the assimilation model could be due to the assumed extended period of isolation in the model being wrong and not be due to the admixture component. The small confidence interval they obtained for the admixture rate does not discriminate between these two biological interpretations because this small confidence interval arises directly from their assumption of a period of prior isolation. Thus, these two components of their assimilation model are confounded in their simulations and no clear biological interpretation for the reason for rejecting the assimilation model is possible.
In contrast, multilocus NCPA can test individual features of complex models. For example, with 95% confidence, archaic Eurasians and archaic Africans have been exchanging genes under an isolation by distance model with no significant interruption for the past 1.46 million years (Templeton 2005). Phrased as a null hypothesis, this means that we can reject the null hypothesis of isolation between these archaic human populations over the past 1.46 million years at the 5% level of significance. The statistical framework of multilocus NCPA is easily extended to test the null hypothesis of no gene flow (isolation) between two geographical areas in an arbitrary interval of time, say l to u. Let ti be the random variable describing the possible times of inferred gene flow from locus i between two geographical areas of interest, Ti the estimated mean time of an NCPA inference of gene flow from locus i between the two areas, and ki the average pairwise mutational divergence between haplotypes that arose since Ti at locus at locus i. Then, using the gamma distribution described in Templeton (for more details see Templeton 2002b, 2004a), the probability of no gene flow from locus i in the interval l to u is:
| 1 |
Under the null hypothesis of isolation of the two areas in the time interval l to u, the probability of no gene flow is 1. Hence, if j is the number of loci that yield an inference of gene flow between the two areas of interest, the likelihood ratio test (LRT) of the null hypothesis of isolation between the two areas in the time interval l to u is:
| 2 |
with the degrees of freedom being j because the null hypothesis that all loci yield a probability of 1 is of zero dimension.
The minimum interval of isolation in the assimilation model of Fagundes et al. (2007) is between 4,000 generations (80,000 years ago) to 32,000 generations (640,000 years ago) from their supplementary Table 7. As this minimum interval is fully contained within their broader interval, rejection of the null hypothesis for the minimum interval automatically implies rejection of isolation in the broader time interval, so multiple testing is not necessary. With the data given in Templeton (Templeton 2005, 2007b) on 18 cross-validated inferences of gene flow between African and Eurasian populations during the Pleistocene, the null hypothesis of complete genetic isolation between Africans and Eurasians during this time interval is rejected with a likelihood ratio test value of 30.02 and 18 degrees of freedom, yielding a probability level of 0.0094. Hence, the isolation component of their assimilation model has been strongly falsified by testing it as a null hypothesis.
The second component of their confounded assimilation model is admixture with the expanding out-of-Africa population. The null hypothesis of no admixture (replacement) is rejected with a probability level of less than 10−17 (Templeton 2005). Other recent studies (Cox et al. 2008; Eswaran et al. 2005; Garrigan& Kingan 2007; Garrigan et al. 2005; Plagnol& Wall 2006) also report evidence of admixture. Hence, the part of the assimilation model of Fagundes et al. (2007) that is wrong (and that is also discrepant with observed autosomal coalescence times) is the part concerning prior isolation of small populations of archaic Africans and Europeans and not the admixture component. The ability of the robust hypothesis-testing framework of NCPA to separate out different phylogeographic components is a great advantage over ABC that produces inferences that have no clear biological interpretations when complex phylogeographic hypotheses are being simulated.
Even assumptions not directly related to the phylogeographic model can influence phylogeographic conclusions. Programs such as SIMCOAL, used by Fagundes et al. (2007), specifically exclude mutation models that depend on the nucleotide composition of the sequence. However, mutation in humans is highly non-random, and all of this non-randomness depends upon the nucleotide composition (Templeton et al. 2000a and references therein). Hence, the mutational models used in SIMCOAL are known to be unrealistic for human nuclear DNA. Unrealistic mutation models in turn can influence phylogeographic inference. For example, Palsbøll et al. (2004) surveyed mitochondrial DNA (mtDNA) in fin whales (Balaenoptera physalus) from the Atlantic coast off Spain and the Mediterranean Sea in order to test two alternative hypotheses: recent divergence with no gene flow versus recurrent gene flow. They discovered that if they used a finite sites mutation model, they inferred recurrent gene flow, but when they used an unrealistic infinite sites mutation model, they inferred the recent divergence model. Until there is a more general assessment of the problems documented by Palsbøll et al. (2004), it is unwise to accept any inferences from simulation programs such as SIMCOAL that use unrealistic models of mutation. In contrast, NCPA does not have to simulate a mutational process but simply uses the mutations that are inferred from statistical parsimony. Statistical parsimony, the first step in the cladistic analysis of haplotype trees, has proven to be a powerful analytical tool in revealing these non-random patterns of mutation (Templeton et al. 2000a) precisely because it is robust to complex models of mutation. The phylogenetic ambiguities that non-random mutagenesis creates can then be incorporated into tree association tests (Brisson et al. 2005; Templeton et al. 2005; Templeton& Sing 1993).
Sampling Error
The field of statistics focuses upon the error induced by sampling from a population of inference. In NCPA, the sampling distributions of the primary statistics are estimated under the null hypothesis by a random permutation procedure. The statistical properties of permutational distributions have been long established in statistics (Edgington 1986), and NCPA implements the permutational procedure in such a manner as to capture both sources of sampling error; a finite number of individuals being sampled, and a finite number of locations being sampled. Other suggested permutational procedures (Petit 2008; Petit& Grivet 2002) do not capture all of the sampling error under the null hypothesis, and such flawed procedures can create misleading artifacts (Templeton 2002a). Hence, sampling error is fully and appropriately taken into account by NCPA.
Another source of error in NCPA is the randomness associated with the coalescent process itself and the associated accumulation of mutations, which will be called evolutionary stochasticity in this paper. The multilocus NCPA explicitly takes evolutionary stochasticity into account through the cross-validation procedure, and variances induced by both the coalescent and mutational processes are explicitly incorporated into the likelihood ratio tests used for cross-validation and for testing specific phylogeographic hypotheses.
There is a misconception that the single-locus NCPA ignores evolutionary stochasticity and equates haplotype trees to population trees (Knowles& Maddison 2002). However, the nested design of NCPA insures that most inferences focus upon just one branch and haplotypes near that branch. This local inference structure gives NCPA great robustness to evolutionary stochasticity and ensures that inferences are not based upon the overall topology of the haplotype tree. Figure 1 illustrates this robustness with respect to five haplotype trees (mtDNA, Y-DNA, and three X-linked regions) from African savanna, African forest, and Asian elephants (Roca et al. 2005). NCPA was performed on the five African haplotype trees, using the Asian haplotypes as the outgroup. In all five cases, a single fragmentation event was inferred at the locations shown by the arrows in each haplotype tree in Figure 1. In all five cases, this fragmentation event primarily separated the savanna from the forest populations. The null hypothesis that all five genes are detecting a single fragmentation event is accepted (the log-likelihood ratio test is 1.497 with 4 degrees of freedom, yielding a probability tail value of 0.8272). Hence, the same inference is cross-validated from five different haplotype trees. Note from Figure 1 that each of the five trees differs in branch lengths and in the topological positions of the three taxa (Figure 1). Only BGN corresponds to a clean species tree. Hence, four of the five trees were influenced by lineage sorting and/or introgression, yet the inferences from single-locus NCPA were completely robust to these potential complications, and the multilocus cross-validation procedure confirms this robustness through a likelihood ratio test. This example also illustrates that NCAP inference does not depend upon equating a haplotype tree to a population tree as all five haplotype trees in this case would yield different population trees under such an equation. Indeed, NCPA does not even assume that a population tree exists at all, as turned out to be the case in human evolution that is characterized by a trellis of genetic interchanges rather than a population tree (Templeton 2005).
In ABC there is no null hypothesis, which complicates the computation of sampling error since there is no single statistical model under which to evaluate sampling error. Fortunately, Ewens (1983) clarified the sampling issues in situations such as this. He pointed out that there are two basic categories of statistics used in population genetics: “long-term” statistics whose sampling error includes both the randomness of the evolutionary process that created the current population (evolutionary stochasticity) and the error associated with a finite sample from the current population; and “current generation” statistics whose sampling error includes only that associated with a finite sample from the current generation. The errors associated with these two types of statistics are shown pictorially in Figure 2. This clarification of sources of sampling error led to a whole new generation of statistics in population genetics, such as the Tajima D statistic (Tajima 1989). Tajima's D statistic, and many subsequent ones, can be expressed generically as:
| 3 |
where Scg is the current generation statistic, Slt is the long-term statistic, and ||•|| designates some sort of normalization operation. This is the same class of statistics used in the ABC method (Beaumont et al. 2002), which is based on ||s’-s|| where s is the observed value from the sample from the current generation and s’ is the value obtained from a long-term coalescent simulation that yields the current generation followed by a sampling scheme identical to that used to obtain s. Tajima (1989), like Ewens (1983), noted that to test the evolutionary model being invoked for the Slt statistic, the goodness of fit statistic in equation 3 must take into account all sources of sampling error in both the Scg and Slt statistics (Figure 2). Hence, Tajima (1989) calculated the evolutionary stochasticity and current generation sampling error of Slt and the current generation sampling error of Scg. It is at this point that a major discrepancy appears between ABC and Tajima's use of equation 3. In ABC, s’ is the long-term statistic, and the contribution of evolutionary stochasticity and current generation sampling error is taken into account via the computer simulation. However, s is the current generation statistic, and it is treated as a fixed constant in the ABC methodology, thereby violating the known sources of error made explicit by Ewens (1983) and that are incorporated into population genetic statistics such as Tajima's D statistic (Tajima 1989). The impact of treating s as a fixed constant is to increase statistical power as an artifact. This increase in pseudo-power can be quite substantial for small sample sizes (see equation 29 in Tajima, 1989), and explains why great statistical resolution is claimed for the ABC method based on miniscule sample sizes (Fagundes et al., 2007). Ignoring the sampling error of s undermines the statistical validity of all inferences made by the ABC method.
Figure 2.
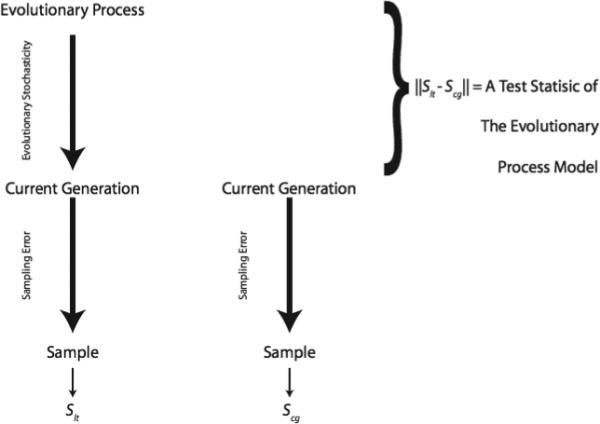
A diagram of the sampling considerations made explicit by Ewens (1983) for long-term statistics (Slt) and current generation statistics (Scg). A contrast of these two types of statistics will focus its power on the evolutionary model used to generate the long-term statistic only if both types of statistics include the impact of sampling error from the current generation.
The pseudo-power achieved by ignoring sampling error in s also undermines the primary method of validation of the ABC: the analysis of simulated populations. Validating a simulation method of inference with simulations hides the problem of unrealistic assumptions, as these assumptions are typically shared by both the analytical method and by the simulated data sets. The problems of weak inference and of ad hoc interpretative sets are ignored because the true model is typically included in the interpretative set. By including the true model in the interpretative set, the pseudo-power created by ignoring sampling error will cause the ABC method to appear to perform well with these simulated data sets. Unfortunately, when the truth is not fully known (i.e., real data), this pseudo-power can generate many false positives and misleading inferences.
Full Distributions vs. local probabilities
The random permutation procedure of NCPA generates the full sampling distribution under the null hypothesis of no geographical associations. In contrast, the ABC method only generates local probabilities in the vicinity of the observed statistics (||s’-s|| ≤ δ where δ is a number that determines the level of acceptable goodness of fit). Bayesian inference is based upon having the posterior distribution, not just local probabilities. The local behavior of the posterior distributions can sometimes be very misleading, as shown in Figure 3. This figure shows a case in which there is a single, one-dimensional statistic, and ABC concentrates its inference in a small region around the observed value. The complete posterior distributions for three models are shown in Figure 3. Consider first models I and II. The observed statistic lies in the tails of both posterior distributions. This is probably a common situation. For example, Ray et al (2005) simulated the out-of-Africa replacement model and other models of human evolution, but they found that even their best fitting model only explained about 10% of the variation. Hence, the observed statistic falling on the tail of the posterior is not unlikely. Note that in Figure 3, model I has a higher relative probability (the area under the curve in the local area indicated around the observed statistic) than model II. Yet, the observed statistic is actually closer to the higher values where model II places most of its probability mass than to the lower values where model I places most of its probability mass. Hence, if one had the entire posterior distributions, the conclusion about which of these two models was the better fitting might well be reversed.
However, it is model III in Figure 3 that is the clear winner, having much more local probability mass than either models I or II. Note that model III has a flat posterior. This can occur for two reasons. First, it may be that model III is completely uninformative relative to the calculated statistic; that is, the data are irrelevant to this model. Alternatively, model III may be over-determined such that it has so many parameters relative to the sample size that it can fit equally well to any observed outcome. As Figure 3 illustrates, the ABC method cannot distinguish between a truly good fitting model, an uninformative model, or an over-determined model. This is the danger of using only local relative probabilities rather than true posterior distributions.
Sample Size
NCPA is computationally efficient, so even large sample sizes can be handled on a desk top computer. For example, the multilocus NCPA of human evolution included data sets with up to 42 locations and sample sizes of 2320 individuals (Templeton 2005), and this is not the limit for NCPA. Because ABC must simulate every population in a complex model, ABC has severe constraints on sample size. For example, Fagundes et al. (2007) had the genetic data on all 50 loci for five populations: 10 sub-Saharan Africans, 10 Europeans, 2 East Indians, 8 East Asians, and 12 American Indians. However, the data from the European and East Indian Eurasian populations were “excluded from the analysis to avoid incorporating additional parameters in our scenarios” (SI text of Fagundes et al. 2007). The inability of ABC to deal with just 5 locations and a total sample size of 42 individuals in this case is a serious defect for any phylogeographic tool. As shown by the inference key for NCPA, many types of phylogeographic inference cannot be made if few sites are sampled, such as distinguishing fragmentation from isolation-by-distance.
There is no obvious statistical rationale for the populations actually analyzed by Fagundes et al. (2007). Given that the American portion of their simulations is identical in all three models, the 12 American Indians are irrelevant to discriminating among three models of human evolution that they tested. The three models differ only in their relationship between African and Eurasian populations, yet the majority of their Eurasian sample was excluded from the analysis. The 40% of their data that is irrelevant to the tested models flattens the posterior. The informative subset of the data consist of just two geographical regions and 18 individuals. Because of the geographical hierarchy in sampling, the degrees of freedom in the informative data subset is bounded between 2 and 18, making over-determination of their models a real possibility as the number of parameters varied from 10 to 18 after excluding the parameters related to the Americas.
Approximate Probabilities in NCPA versus ABC
NCPA approximates the null distribution via a well characterized random permutation procedure (Edgington 1986). The convergence of the approximation depends upon the number of random permutations performed. The default in NCPA is 1,000 random permutations, which is adequate for most statistical inference. However, if any user wants more precision in the approximation, it is easily manipulated simply by increasing the number of permutations.
ABC approximates its local posterior probabilities via random computer simulations of complex scenarios. The convergence of this approximation depends upon the parameter δ (Beaumont et al. 2002). However, Beaumont et al. (2002) do not examine the convergence properties and only give ad hoc, heuristic guidelines in choosing δ. They also do not investigate the impact of sample size on the convergence of these probability measures. An Euclidian space is defined for the observed statistics even though many of the statistics used do not naturally fall into a Euclidian space, such as the number of segregating sites. As shown by Billingsley (1968), convergence of probability measures depends in part on the type of space being used, and it is not clear what mixing statistics that naturally fall into different types of spaces would do to the convergence properties. Finally, a spherical Euclidian space is assumed. Spherical spaces should be invariant to rigid rotations and reflections of their axes, yet the highly correlated nature of the statistics being used patently violates the assumption of sphericity. The impact of all of these factors upon the convergence properties of these probability measures is not addressed by Beaumont et al. (2002).
Approximations are common in statistics, but they should only be used when the limits of the approximation are known. For example, Table 1 gives a hypothetical two-by-two contingency table. One can test the null hypothesis of homogeneity in such a table with a chi-square goodness of fit, which approximates the null distribution under some conditions. Doing so yields a chi-square of 4 with 1 degree of freedom, which is significant at the 0.05 level. However, the bottom row has too few observations in this case for the chi-square approximation to be valid. Instead, Fisher's exact test should be used in this situation, and with this test there is no significant departure from homogeneity at the 0.05 level. The ABC method, like any approximation in statistics, should never be used unless the user is confident that their choices for δ, sample size, and statistics allow the approximation to be valid. This was not done by Fagundes et al. (2007).
Table 1.
A hypothetical two-by-two contingency table.
| A1 |
A2 |
|
|---|---|---|
| B1 | 52 | 20 |
| B2 | 1 | 3 |
Dimensionality and Comeasurability
How well a model fits a given data set depends not only upon the extent to which the model captures reality, but also upon the dimensionality of the model relative to the size and structure of the sample. For example, a Hardy-Weinberg model can always fit exactly the phenotypic data from a one-locus, two allele, dominant allele model regardless of whether or not the population is in Hardy-Weinberg. Dominance means that there is only one degree of freedom available from the data, and one degree of freedom is used to estimate the allele frequency under Hardy-Weinberg. The resulting goodness of fit statistic indicates a perfect fit, but the statistic has zero degrees of freedom. Hence, the perfect fit of the Hardy-Weinberg model to a dominant allele model is meaningless statistically. As this example illustrates, the goodness of fit of a model and its statistical support can be very different.
NCPA builds up its inferences from simple, one-dimensional tests at the single locus level. The cross validation statistics and the tests of specific phylogeographic hypotheses can be of higher dimension in NCPA. All of these higher dimension tests are nested in that the null hypothesis can be regarded as a lower-dimension special case relative to alternative hypotheses. This is important because the statistical theory for testing nested hypotheses is well developed and straightforward whereas it is far more difficult to compare hypotheses that are not nested. Moreover, the parameters in the alternative models all play a similar role with respect to their respective probability distributions. As a consequence of nesting and comparable parameterization, the degrees of freedom in the NCPA multilocus tests are easy to calculate, so over-determined models can be avoided. For example, in testing the out-of-Africa replacement model with likelihood ratio test, the degrees of freedom were 17, indicating that much information was available in the multilocus data set to test this hypothesis.
Unlike NCPA, ABC invokes complex phylogeographic models at the onset. ABC then uses a goodness-of-fit criteria upon these models. Indeed, ||s’-s|| is a generalized goodness of fit statistic that includes the standard chi-square statistic of goodness of fit. The goodness-of-fit nature of the ABC is further accentuated by the application of the Epanechnikov kernel and local regression (Beaumont et al. 2002). How close a simulated model will approximate the observed statistics will depend in part upon the number and nature of the parameters in the model and upon the size and structure of the data set. This information is needed in order to interpret the goodness of fit statistic. This is not only true of likelihood ratio tests, but of Bayesian procedures as well (Schwarz 1978). The dimensionality of the models used in ABC has not been determined in any application that I have seen, and certainly not in Fagundes et al. (2007). Because the models are complex, determining dimensionality is not just a simple task of counting up the number of parameters, as different parameters influence the models in qualitatively different fashions and interact with one another in the simulation. Further complicating dimensionality of test statistics is the fact that the models in ABC are often not nested, and one model may contain parameters that do not have analogues in the other models and vice versa. Finally, the data are often sampled in a hierarchical fashion in phylogeographic studies, making the calculation of the available degrees of freedom difficult.
Knowing hypothesis dimensionality is critical for valid statistical inference. For example, suppose two models were being evaluated with a traditional chi-square goodness of fit statistic, and model I yields a chi-square statistic of 5 and model II yields a chi-square statistic of 10. Which is the better fitting model? Since the chi-square statistic decreases in value as the model fits better and better, it would be tempting to say that model I is the better fitting model as its chi-square goodness of fit statistic is smaller. However, suppose that model I has 1 degree of freedom and model II has 5. Using this information, we can transform the chi-square statistics into tail probabilities, yielding a probability of 0.025 for model I and 0.075 for model II. Hence, model II actually fits the data better than model I.
This hypothetical example illustrates the importance of a property called comeasurability. Comeasurability requires an absolute ability to say if A > B, A=B, or A < B. Goodness of fit statistics from the class ||s’-s|| do not have the property of comeasurability, as illustrated by the chi-square example above. For statistical inference, it is critical to have a metric that is comeasurable, and a probability measure is one such entity. That is why the chi-square goodness of fit statistics had to be transformed into tail probabilities in order to compare the fits of models I and II. Indeed, much of statistical theory is devoted to transforming statistics that are not inherently comeasurable (chi-squares, t-tests, likelihood ratios, mean squares, least-squares, etc.) into comeasurable probability statements.
The posterior probabilities in ABC are constructed by using a numerator that is a function of the goodness of fit measure ||s’-s|| for a particular model. The numerator is then divided by a denominator that ensures that the “probabilities” sum to one across the finite set of hypotheses (see equation 9 in Beaumont et al. 2002). Beaumont et al. (2002) make no adjustments for the dimensionality of these hypotheses, violating the Schwarz (1978) proposition, nor do they distinguish between nested and non-nested alternative models. As a result, the numerators are not comeasurable across hypotheses, and the denominators are sums of non-comeasurable entities. Hence, the “posterior probabilities” that emerge from ABC are not comeasureable. This means that it is mathematically impossible for them to be probabilities.
Fagundes et al. (2007) assign an ABC “posterior probability” of 0.781 to the out-of-Africa replacement model. Because it is mathematically impossible for this number to be a probability, the out-of-Africa replacement model is not necessarily the most probable model out of the three that they considered. Indeed, the number 0.781 has no meaningful statistical interpretation. Thus, the final product of the ABC analysis are numbers that are devoid of statistical meaning. The ABC method is not capable of even weak statistical inference.
Discussion
Table 2 summarizes the points made in this paper about the statistical properties of multilocus NCPA and ABC. As can be seen, ABC has multiple statistical flaws and does not yield true probabilities. In contrast, multilocus NCPA provides a framework for hard inference based upon well-established statistical procedures such as permutation testing and likelihood ratios. There has been much criticism of NCPA starting in 2002. Some of this criticism is based on factual errors; such as the misrepresentation that NCPA equates haplotype trees to population trees, or that nested clade analysis cannot analyze data other than geographical data. Other criticisms had validity; such as the interpretations of single-locus NCPA were not phrased as testable null hypothesis or that the false positive rate was high. These flaws of single locus NCPA were addressed and solved by multilocus NCPA. Consequently, the criticisms of Knowles and Maddison (2002), Panchal and Beaumont (2007), Petit (2008), and Beaumont and Panchal (2008) are irrelevant to multilocus NCPA. As shown in this paper, multilocus NCPA is a robust and powerful method of making hard phylogeographic inference and does not suffer from the limitations attributed to single-locus NCPA.
Table 2.
Properties of multilocus Nested Clade Analysis versus Approximate Bayesian Computation.
| Property | NCA | ABC |
|---|---|---|
| Genetic Data Used | Haplotype Trees | Broad Array of Genetic Data |
| Data Analyzable | Geographic, Phenotypic, Ecological, Interspecific, etc. | Geographic, Ecological, Interspecific, etc. |
| Nature of Inference | Strong | Weak |
| Interpretative Criteria | Explicit, A priori, Universal | Explicit, Ad hoc, Case Specific |
| False Positives | False Positive Rate Estimable; False Positives Reduced via Cross-Validation | False Positive Rate Not Estimable; No Mechanism for Correcting for False Positives |
| A Priori Phylogeographic Model | No: Allows Discovery of Unexpected | Yes: Inference Limited to Finite Set of a priori Alternatives |
| Nature of Inferred Phylogeographic Model | Built-up From Simple Components, Each with Explicit Statistical Support | Full Models Specified a priori, Resulting in Confoundment When Model Has Several Components |
| Mechanics of Inference | Interpretive Key Followed By Phrasing as Null Hypotheses Tested With Likelihood Ratios | Simulation Requiring Multiple Parameters: Model and Parameter Values Confounded |
| Sampling Error | Incorporates Errors Due to Tree Inference, No. of Locations, No. of Individuals, and Evolutionary Stochasticity | Incorporates Sampling Error and Evolutionary Stochasiticy in Simulations; Ignores Error In Current Generation Statistics |
| Probability Distributions and Probablities | Simulates Full Sampling Distribution; Likelihood Ratios Based on Full Distributions | Local Probabilities, Obscuring Good-Fitting, Irrelevant, and Over-Determined Models |
| Sample Size | Handles Large Numbers of Locations and Individuals | Severely Restricted on the Number of Locations Analyzable |
| Convergence | Well Defined | Unknown |
| Dimensionality of Tests | Well Defined | Ignored |
| Final Test Product | Tail Probability of Null Hypothesis | A Non-Comeasurable Fit Metric With No Probabilistic Meaning |
Because of its multiple flaws, ABC should not be used for hypothesis testing. This does not mean that simulation approaches have no role in statistical phylogeography. The strength of NCPA is to falsify hypotheses and to build up complex phylogeographic models from simple components without any need for prior knowledge. NCPA achieves its robustness in testing hypotheses by taking a non-parametric approach, but this also means that NCPA will not yield much insight into the details of the emerging phylogeographic models. For example, isolation-by-distance may be inferred, but there is no estimation of the parameters of an isolation-by-distance model. A range expansion might be inferred, but there is no insight into the demographic details of that expansion through NCPA. Moreover, NCPA is limited to haplotype data from DNA regions with little to no recombination, but other types of data may have considerable phylogeographic information as well. Hence, the best phylogeographic analyses are those that use NCPA or some other statistically valid procedure to outline the basic phylogeographic model, followed by the use of simulation techniques to estimate phylogeographic and gene flow parameters and to incorporate additional information from other types of data (for some examples, see Brown& Stepien 2008; Garrick et al. 2008; Garrick et al. 2007; Strasburg et al. 2007). Of particular relevance is the work of Gifford and Larson (2008) who used multilocus NCPA as their primary hypothesis testing tool and coalescent-based simulations for some parameter estimation. Hence, NCPA and simulation approaches are not so much alternative techniques as they are complementary, and potentially synergistic, techniques. Both add to the statistical toolkit of intraspecific phylogeographers, and both should be used when appropriate.
Acknowledgments
This work was supported in part by NIH grant P50-GM65509. I wish to thank six anonymous reviewers for their useful suggestions and Laurent Excoffier for patiently explaining some of the details of the ABC analysis of human evolution.
Footnotes
Arthur Information Box:
Alan Templeton has both a Master's Degree in Statistics and a Doctoral degree in Human Genetics. This combination of statistical and genetics training has allowed him to develop many innovative statistical methods in human genetics and evolutionary biology, including the statistical procedure of nested clade analysis.
References
- Avise JC, Arnold J, Ball RM, et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annual Review of Ecology and Systematics. 1987;18:489–522. [Google Scholar]
- Avise JC, Lansman RA, Shade RO. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics. 1979;92:279–295. doi: 10.1093/genetics/92.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Panchal M. On the validity of nested clade phylogeographical analysis. Molecular Ecology. 2008;17:2563–2565. doi: 10.1111/j.1365-294X.2008.03786.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Zhang WY, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovici S, Geiger D, Shlush L, Skorecki K, Templeton A. Panel construction for mapping in admixed populations via expected mutual information. Genome Research. 2008;18:661–667. doi: 10.1101/gr.073148.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley P. Convergence of Probability Measures. John Wiley & Sons, Inc.; New York: 1968. [Google Scholar]
- Brisson JA, De Toni DC, Duncan I, Templeton AR. Abdominal pigmentation variation in Drosophila polymorpha: Geographic variation in the trait, and underlying phylogeography. Evolution. 2005;59:1046–1059. [PubMed] [Google Scholar]
- Brown JE, Stepien CA. Ancient divisions, recent expansions: phylogeography and population genetics of the round goby Apollonia melanostoma. Molecular Ecology. 2008;17:2598–2615. doi: 10.1111/j.1365-294X.2008.03777.x. [DOI] [PubMed] [Google Scholar]
- Cox MP, Mendez FL, Karafet TM, et al. Testing for Archaic Hominin Admixture on the X Chromosome: Model Likelihoods for the Modern Human RRM2P4 Region From Summaries of Genealogical Topology Under the Structured Coalescent. Genetics. 2008;178:427–437. doi: 10.1534/genetics.107.080432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington ES. Randomization Tests. 2nd edn. Marcel Dekker; New York and Basel: 1986. [Google Scholar]
- Eswaran V, Harpending H, Rogers AR. Genomics refutes an exclusively African origin of humans. Journal of Human Evolution. 2005;49:1–18. doi: 10.1016/j.jhevol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Ewens WJ. The role of models in the analysis of molecular genetic data, with particular reference to restriction fragment data. In: Weir BS, editor. Statistical Analysis of DNA Sequence Data. Marcel Dekker; New York: 1983. pp. 45–73. [Google Scholar]
- Fagundes NJR, Ray N, Beaumont M, et al. Statistical evaluation of alternative models of human evolution. Proceedings of the National Academy of Sciences. 2007;104:17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick RC, Rowell DM, Simmons CS, Hillis DM, Sunnucks P. Fine-scale phylogeographic congruence despite demographic incongruence in two low-mobility saproxylic springtails. Evolution. 2008;62:1103–1118. doi: 10.1111/j.1558-5646.2008.00349.x. [DOI] [PubMed] [Google Scholar]
- Garrick RC, Sands CJ, Rowell DM, Hillis DM, Sunnucks P. Catchments catch all: long-term population history of a giant springtail from the southeast Australian highlands – a multigene approach. Molecular Ecology. 2007;16:1865–1882. doi: 10.1111/j.1365-294X.2006.03165.x. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB. Archaic human admixture: A view from the genome. Current Anthropology. 2007;48:895–902. [Google Scholar]
- Garrigan D, Mobasher Z, Severson T, Wilder JA, Hammer MF. Evidence for Archaic Asian Ancestry on the Human X Chromosome. Mol Biol Evol. 2005;22:189–192. doi: 10.1093/molbev/msi013. [DOI] [PubMed] [Google Scholar]
- Gifford ME, Larson A. In situ genetic differentiation in a Hispaniolan lizard (Ameiva chrysolaema): A multilocus perspective. Molecular Phylogenetics and Evolution. 2008;49:277–291. doi: 10.1016/j.ympev.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Knowles LL. The burgeoning field of statistical phylogeography. J Evolution Biol. 2004;17:1–10. doi: 10.1046/j.1420-9101.2003.00644.x. [DOI] [PubMed] [Google Scholar]
- Knowles LL, Maddison WP. Statistical phylogeography. Molecular Ecology. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Lemmon EM. A Likelihood Framework for Estimating Phylogeographic History on a Continuous Landscape. Systematic Biology. 2008;57:544–561. doi: 10.1080/10635150802304761. [DOI] [PubMed] [Google Scholar]
- Palsbøll PJ, Berube M, Aguilar A, Notarbartolo-Di-Sciara G, Nielsen R. Discerning between recurrent gene flow and recent divergence under a finite-site mutation model applied to North Atlantic and Mediterranean Sea fin whale (Balaenoptera physalus) populations. Evolution. 2004;58:670–675. [PubMed] [Google Scholar]
- Panchal M, Beaumont MA. The automation and evaluation of nested clade phylogeographic analysis. Evolution. 2007;61:1466–1480. doi: 10.1111/j.1558-5646.2007.00124.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ. The coup de grace for the nested clade phylogeographic analysis? Molecular Ecology. 2008;17:516–518. doi: 10.1111/j.1365-294X.2007.03589.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Grivet D. Optimal randomization strategies when testing the existence of a phylogeographic structure. Genetics. 2002;161:469–471. doi: 10.1093/genetics/161.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Wall JD. Possible Ancestral Structure in Human Populations. PLoS Genetics. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper KR. The logic of scientific discovery. Hutchinson; London: 1959. [Google Scholar]
- Pritchard JK, Seielstad MT, Perez-Lezaan A, Feldman MW. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Molecular Biology and Evolution. 1999;16:1791–1978. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- Ray N, Currat M, Berthier P, Excoffier L. Recovering the geographic origin of early modern humans by realistic and spatially explicit simulations. Genome Research. 2005;15:1161–1167. doi: 10.1101/gr.3708505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca AL, Georgiadis N, O'Brien SJ. Cytonuclear genomic dissociation in African elephant species. Nature Genetics. 2005;37:96–100. doi: 10.1038/ng1485. [DOI] [PubMed] [Google Scholar]
- Schwarz G. Estimating the Dimension of a Model. The Annals of Statistics. 1978;6:461–464. [Google Scholar]
- Strasburg J, Kearney M, Moritz C, Templeton A. Combining phylogeography with distribution modeling: multiple pleistocene range expansions in a parthenogenetic gecko from the Australian arid zone. PLoS ONE. 2007;2:e760. doi: 10.1371/journal.pone.0000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Lee S-H, Satta Y. Testing Multiregionality of Modern Human Origins. Mol Biol Evol. 2001;18:172–183. doi: 10.1093/oxfordjournals.molbev.a003791. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Molecular Ecology. 1998;7:381–397. doi: 10.1046/j.1365-294x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Using phylogeographic analyses of gene trees to test species status and processes. Molecular Ecology. 2001;10:779–791. doi: 10.1046/j.1365-294x.2001.01199.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. “Optimal” randomization strategies when testing the existence of a phylogeographic structure: A reply to Petit and Grivet. Genetics. 2002a;161:473–475. doi: 10.1093/genetics/161.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Out of Africa again and again. Nature. 2002b;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- Templeton AR. A maximum likelihood framework for cross validation of phylogeographic hypotheses. In: Wasser SP, editor. Evolutionary Theory and Processes: Modern Horizons. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004a. pp. 209–230. [Google Scholar]
- Templeton AR. Statistical phylogeography: methods of evaluating and minimizing inference errors. Molecular Ecology. 2004b;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Haplotype trees and modern human origins. Yearbook of Physical Anthropology. 2005;48:33–59. doi: 10.1002/ajpa.20351. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Perspective: Genetics and recent human evolution. Evolution. 2007a;61:1507–1519. doi: 10.1111/j.1558-5646.2007.00164.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. Population biology and population genetics of Pleistocene Hominins. In: Henke W, Tattersall I, editors. Handbook of Palaeoanthropology. Springer-Verlag; Berlin: 2007b. pp. 1825–1859. [Google Scholar]
- Templeton AR. Nested clade analysis: an extensively validated method for strong phylogeographic inference. Molecular Ecology. 2008;17:1877–1880. doi: 10.1111/j.1365-294X.2008.03731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Boerwinkle E, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of Alcohol Dehydrogenase activity in Drosophila. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Clark AG, Weiss KM, et al. Recombinational and mutational hotspots within the human Lipoprotein Lipase gene. American Journal of Human Genetics. 2000a;66:69–83. doi: 10.1086/302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Maskas SD, Cruzan MB. Gene Trees: A Powerful Tool for Exploring the Evolutionary Biology of Species and Speciation. Plant Species Biology. 2000b;15:211–222. [Google Scholar]
- Templeton AR, Maxwell T, Posada D, et al. Tree scanning: a method for using haplotype trees In genotype/phenotype association studies. Genetics. 2005;169:441–453. doi: 10.1534/genetics.104.030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Routman E, Phillips C. Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the Tiger Salamander, Ambystoma tigrinum. Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics. 1993;134:659–669. doi: 10.1093/genetics/134.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]


