Abstract
Background
In humans, thermal cutaneous injury represents a serious traumatic event that induces a host of dynamic alterations. Unfortunately the molecular mechanisms that underlie these serious perturbations remain poorly understood. We applied a global analysis method to identify dynamically changing proteins within the burn environment, which could eventually become biomarkers or targets for treatment.
Materials and Methods
Protein extracts of normal/unwounded skin and burn wounds were assayed by 2D difference gel electrophoresis (DIGE), a proteomic technology by which abundance levels of intact proteins (including isoforms) were simultaneously quantified from multiple samples with statistical confidence. Through unsupervised multivariate principal component analysis, protein expression patterns from individual samples were appropriately clustered into their correct temporal healing periods grouped into postburn periods of 1–3 days, 4–6 days or 7–10 days after injury. Forty-six proteins were subsequently selected for identification by matrix assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS).
Results
Proteins identified with differential temporal patterns of expression included predictable cytoskeletal proteins such as vimentin, and keratins 1, 5, 6, 16, and 17. Other candidate proteins with potential involvement in healing included HSP90, members of the serpin family (Serpin B1, SCCA1 & 2), haptoglobin, gelsolin, eIF4A1, IQGAP1, and TCTP.
Conclusions
We have utilized the combined technique, DIGE/MS, to capture new insights into cutaneous responses to burn trauma and subsequent processes of early wound healing in humans. This pilot study provides a proteomic snapshot of temporal events that can be used to weave together the interconnected processes that define the response to serious cutaneous injury.
Keywords: wound healing, proteomics, 2D-DIGE, burns, skin
Introduction
Cutaneous injury following thermal trauma induces a host of molecular changes both locally within the burned skin area and its adjacent margins as well as systemically. In the acute phase after injury, oxidative stress ensues, heat shock proteins are unleashed, and the inflammatory cascade is initiated resulting in an inevitable progression in wound depth (1–3). This acute phase quickly gives rise to inflammatory influx, cellular responses that are marked by cellular migration and proliferation, as well as the production of extracellular matrix molecules. In human burn wounds, this complex series of events typically sets the foundation for undesirable and disfiguring hypertrophic scar formation (4–6). For the severely burned patient, the burn trauma often leads to life-threatening infections, prolonged illness and even death. Fortunately, the advent of better anti-microbial agents, biologic and conventional dressings, and control of infection has drastically reduced the morbidity and mortality rate for the annual 1.25 million burn patients treated in the United States (7, 8). Nevertheless, only incremental progress has been realized largely due to the slow progress directed towards discovering the basic biological changes that drive the acute responses and wound healing processes in humans.
Previous strategies for understanding wound repair mechanisms have largely involved reductionist techniques based on painstaking evaluations of either a single or a few cytokines per study. This stepwise and piecemeal accumulation of data has undoubtedly hindered the development of new burn therapies. Although global approaches such as microarrays are beginning to preview patterns in gene transcripts, this work has not yet extended into global protein analysis. Proteomics can provide a portrait of translational regulation of expression, post-translational expression and changes in steady-state protein levels. Previous proteomic studies in the skin have established a framework of skin or keratinocyte specific proteins associated with normal homeostasis (Huang, 2004). To date, proteome profiles have also emerged for the aging process (Gromov, 2003), and for response to minor stresses induced by UV irradiation (9).
The present study was conducted to test our hypothesis that proteomic analysis could identify unique protein profiles associated with different phases of wound healing. Initially a 2D-DIGE approach, with a pooled-sample internal standard was used to analyze changes in protein abundance between samplings from 3 groups of burn patients at different time points in the healing process alongside a normal/unwounded group. Quantitative measurements of protein abundance changes were obtained with statistical confidence (Student’s t-test, ANOVA). Unsupervised principal component analysis confirmed that a subset of statistically-significant abundance changes could collectively classify the burn or control samples into appropriate classes (rather than occurring stochastically), the classes being normal/unwounded, post burn days (PBD) 0–3 (Early), 4–7 (Middle), and 8–10 (Late). Forty-six of these proteins were unambiguously identified through a combination of matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry, TOF-TOF tandem mass spectrometry, and database interrogation that specified 20 unique proteins including modified isoforms. Our goal was to identify changing target proteins during cutaneous wound repair following substantial thermal injury with future therapeutic value.
Materials and Methods
Sample Collection and Isolation
The study was approved by Vanderbilt University Medical Center’s Institutional Review Board. Informed consent was obtained from the in-patient burn population who were scheduled for excision and skin grafting and from patients undergoing elective surgical procedures. Skin samples were collected from freshly excised skin removed in the operating room. Three patient samples in each of the following categories were used for this analysis: normal/unwounded, PBD 0–3 (Early), PBD 4–7 (Middle), and PBD 8–10 (Late). Tissues containing the healing regions of 1st and 2nd degree burn injuries (partial-thickness) were trimmed into 1cm wide × 2cm long strips that included the healing margins. The more central (full-thickness) regions of damage were discarded. Normal/Unwounded tissues were acquired during elective surgical procedures that included the removal of excess skin. All samples were wrapped in aluminum foil and snap frozen in liquid nitrogen. All tissues were chopped into small pieces at −20°C and thoroughly rinsed in PBS immediately prior to sample lysis.
Protein extraction and quantification
Tissue samples were weighed and 1 mL/gram of lysis solution (9.5M urea, 4% CHAPS, 1X protease inhibitor cocktail, and 2.5% Tributlyphosphine) was used for protein extraction. Samples were then through three cycles of freeze thawing in which the samples were incubated for 10 min in a dry ice bath followed by 10 min at 37°C. Samples were vortexed between cycles and once complete were allowed to sit on ice for 10 min and were then centrifuged at 10,000 × g for 10 min at 4°C. Supernatant were removed and passed through a 26 gauge needle several times. Triplicate aliquots were then quantified by bicinchoninic acid (BCA) protein assay.
Cy-dye labeling and 2D gel electrophoresis and imaging
A pooled internal standard methodology was used in this study (10, 11). Samples were aliquoted, precipitated, and labeled according to previously describe methods using 250 μg protein aliquots from each sample (12). Individual samples were labeled in an alternating fashion, such that not all samples from any one group were ever labeled with the same fluor to control for any dye-specific labeling artifacts. The labeling of each individual sample and its inclusion into the 6-gel matrix was as follows:
| Gel 1 | Gel 2 | Gel 3 | Gel 4 | Gel 5 | Gel 6 | |
| Cy3 | Norm 1 | Norm 2 | Late 1 | Early 2 | Early 3 | Mid 3 |
| Cy5 | Early 1 | Mid 1 | Norm 3 | Mid 2 | Late 2 | Late 3 |
| Cy2 | 12-mix | 12-mix | 12-mix | 12-mix | 12-mix | 12-mix |
This methodology allowed for the direct comparison of individual protein signals between the Cy3- or Cy5 labeled samples relative to the signal for that protein from the Cy2 standard (rather than between two samples directly). Quantitative measurements were always made relative to the cognate signal from the internal standard within each gel, without interference from gel-to-gel variation. Intra-gel ratios were then normalized between gels because the Cy2 standard came from the same labeling mixture, enabling the application of univariate (Student’s t-test and ANOVA) and multivariate (Principal Component Analysis) statistical analyses.
Each mixed set of tripartite-labeled samples was passively rehydrated into 24 cm pH 4–7 immobilized pH gradient (IPG) strips (GE Healthcare, Piscataway, NJ), followed by simultaneous isoelectric focusing, equilibration into the second dimensional loading buffer, and then seating onto 12% homogenous polyacrylamide gels as previously described (12). The second dimension SDS-PAGE was carried out simultaneously on all six gels using standard conditions (13) at less than 1 W per gel for 2 h followed by 20 W per gel until the dye front had reached the bottom using a peltier-cooled DALT II electrophoresis unit (GE Healthcare). Specific Cy dye channels were imaged and then the gels were stained with Sypro Ruby as previously described (12).
DIGE analysis
DeCyder 2D v6.5 software (GE Healthcare) was used for simultaneous comparison of abundance changes across all four sample groups. The DeCyder differential in-gel analysis (DIA) module generated ratios for each protein “spot” by comparing Cy3 and Cy5 signals to the Cy2 control signal. The DeCyder biological variation analysis (BVA) module matched all 18 protein spot maps from the six gels, and to normalize the DIA-generated Cy3:Cy2 and Cy5:Cy2 ratios relative to the Cy2 signals for each resolved feature separately. This enabled the calculation of average abundance changes across all three samples within each test group, and the application of univariate (Student’s t-test, ANOVA) and multivariate (PCA) statistical analyses.
Pair-wise Student’s t-tests and global ANOVA (Analysis of Variants) tests were used to define a subset of proteins that exhibited a trend towards statistically-significant abundance changes (normalized volume ratios) across the samples, where each protein spot was analyzed individually and p-values reflected the probability that the observed change occurred due to random noise. In most cases, changes within the 99th percentile confidence interval (p < 0.01) were chosen for subsequent identification, although in some cases changes up to the 95th percentile were also considered.
The DeCyder extended data analysis (EDA) module was used to perform unsupervised Principal Component Analysis (PCA), which was performed on a subset of the data that exhibited ANOVA p-values within the 95th confidence interval. Each datapoint in the PCA plot represents an individual sample, within which the global expression patterns of these subsets were evaluated. PCA distills a multivariate analysis down to the two most significant sources of variation, PC1 and PC2, between the samples, in this case based on the global expression patterns from the subset of features present in each.
In-gel digestion, mass spectrometry and database interrogation
Proteins of interest were excised, digested, and processed by matrix assisted laser desorption/ionization, time-of-flight mass spectrometry (MALDI-TOF MS)as previously described (12). The MS and MS/MS spectra were collectively used to interrogate sequences present in the SWISS-PROT and NCBInr databases to generate statistically significant candidate identifications using GPS Explorer software (Applied Biosystems) running the MASCOT search algorithm (www.matrixscience.com). Searches were performed without constraining protein molecular weight or isoelectric point, with complete carbamidomethylation of cysteine, partial oxidation of methionine residues, and 1 missed cleavage also allowed in the search parameters. Significant Molecular Weight Search (MOWSE) scores (p < 0.05), number of matched ions, number of matching ions with independent MS/MS matches, percent protein sequence coverage, and correlation of gel region with predicted MW and pI were collectively considered for each protein identification
Results
Experimental Design and Control Conditions for Labeling
Areas of partial thickness second degree injury containing adjacent skin margins were obtained from burn patients at the time of excision prior to skin grafting (N = 9). Normal/Unwounded skin samples (N = 3) were obtained during elective breast reduction surgery. The demographic profile for our sampling of patients is listed in Table 1. The age range of our patient sampling spanned from 6 to 51 years of age with an average age of 25 and a median of 29. Though our sample pool is small, it does reflect a relatively youthful population with a greater capacity for healing and scarring.
Table 1.
Patient Demographics
| Sample | Group/ Post Burn Day | Age | Mean Age | Sex | Total Body Surface Area Burned | Area of Excision |
|---|---|---|---|---|---|---|
| 1 | Normal/ Unwounded | 32 | 29 | F | Not applicable | Breast |
| 2 | Normal/ Unwounded | 39 | F | Not applicable | Breast | |
| 3 | Normal/ Unwounded | 17 | F | Not applicable | Breast | |
| 4 | Early/ 1 | 51 | 29 | F | 80% | Lower Leg |
| 5 | Early/ 3 | 6 | M | 25% | Lower Leg | |
| 6 | Early/ 3 | 29 | M | 15% | Arm | |
| 7 | Middle/ 4 | 16 | 22 | M | 20% | Arm |
| 8 | Middle/ 4 | 27 | F | 3% | Lower Leg | |
| 9 | Middle/ 4 | 24 | M | 11% | Arm | |
| 10 | Late/ 8 | 33 | 20 | M | 20% | Upper Back |
| 11 | Late/ 9 | 9 | F | 4% | Lower leg | |
| 12 | Late/ 10 | 17 | F | 30% | Upper Back |
As described in Materials and Methods, a pooled internal standard methodology was used in this study. Protein extracts were labeled with either Cy3 or Cy5 fluorescent dye and then combined with a Cy2 labeled internal standard containing an equal mix of each of the 12 patient samples. Direct quantitative comparisons were made for each resolved protein feature between a Cy3- or Cy5-labeled sample directly to the cognate signal from the Cy2-labeled internal standard (but not to each other), without interference from gel-to-gel variation. The resulting Cy3:Cy2 and Cy5:Cy2 quantitative ratios were then matched and normalized between the gels using the same signals from the internal standard (as this standard represented one composite mixture that is labeled one time and then aliquoted to each gel). Abundance ratios were then analyzed first globally using unsupervised principal component analysis to assess proper sample assignment and to identify major outliers, and then probed for proteins that were changing significantly between the different samples for subsequent identification using mass spectrometry and database interrogation.
Multivariate Analysis of Protein Profiles from Normal/Unwounded and Wounded Patients
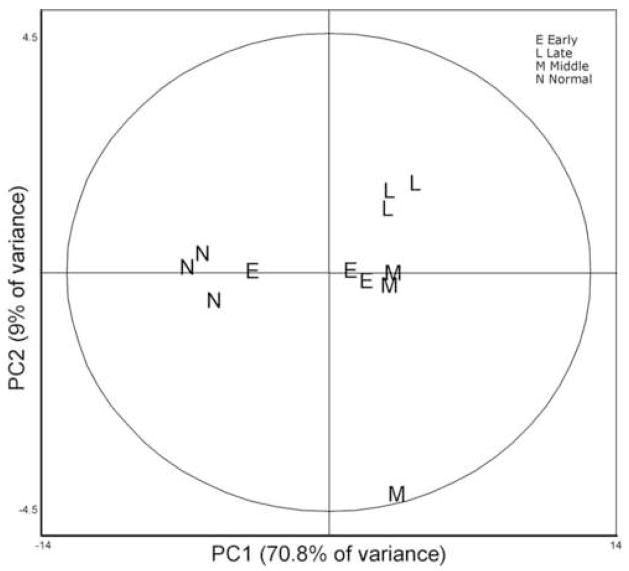
Principal Component Analysis (PCA) was applied using 196 features that displayed significant changes across the four groups (ANOVA p < 0.05). Thus, the patient samples were clustered based on the global expression patterns of these proteins in an unsupervised fashion. PCA reduces the dimensionality of a multi-dimensional analysis to display the two principal components, PC1 and PC2, which distinguish between the two largest sources of variation within the dataset. This form of analysis was useful in determining if any samples were mis-classified based on the extent of burn injury or healing time, or if significant outliers existed in the sample cohort due to person-specific (but not burn-specific) changes.
The 196 expression profiles were considered collectively for each sample, represented as a letter (E = Early Burn, L = Late Burn, M = Middle Burn, N = Normal/Unwounded) indicating the phenotypic class imposed upon the sample, although the analysis was performed in a blinded fashion. In our study analysis, 70.8% of the variation between these 196 protein expression patterns was distinguished by PC1, which clearly segregated the three Normal/Unwounded samples along with one Early sample (Figure 1). A retrospective review of the patient/sample information (Table 1) revealed that this particular sample was collected very early after burn injury (< 24 hours), from the oldest patient in this study, and who was also the only non-survivor. We surmised that insufficient time had elapsed for detectable translational response to injury in this severely-injured, elderly patient. Thus we were able to identify a rational explanation for the appearance of this sample within the cluster of normal samples (Figure 1). Based on this information, this sample was removed from this analysis and the PCA was repeated without it. During this second round of analysis, 231 features from the 11 remaining samples were used to display the significant changes across the four groups. Though more features were selected by the computer on the second PCA analysis, the clustering of the remaining samples did not change (data not shown).
Figure 1.
Unsupervised Principal Component Analysis of individual sample expression maps each consisting of 196 features with ANOVA p < 0.05): A graphical display of the relationship of each sample to one another. No prior sample assignments were made. Data were generated strictly by relative protein abundance data generated from the Biological Variance Analysis. The software shows, without any preconception of sample nature, that the dynamic portions of the sample proteomes are distinct and distinguishable enough to be categorized into a control and 3 temporal groupings.
The second principal component (PC2) distinguished only an additional 9% of the remaining variation, and appeared to segregate the three Late burn samples discretely from the others, with a major outlier found in one of the Middle burn samples. Careful inspection of the patient/sample information for this particular outlier provided no reasonable hypothesis for this segregation and thus at this point can only be attributed to a patient-specific expression pattern. Otherwise, there appeared to be little distinction between samples collected in the Early and Middle timepoints. A much larger patient sampling will be necessary to establish useful patterns from wounds collected during these time periods. Nevertheless, this unsupervised multivariate analysis proved useful in this preliminary study by providing a measure of validation for the arbitrary clustering of wound healing samples into early, middle and slightly later time points of wound repair. This nascent knowledge will form the basis for our future studies which will include a much larger number of patient samples.
In the next phase of our proteomic analysis, MALDI-TOF-MS with database interrogation was applied to the 46 protein features that exhibited statistically-significant abundance changes between any of the 4 groups (assessed using a univariate Student’s t-test). In many cases, p-values within the 99th percentile confidence interval were observed (p < 0.01), but changes within the 95th confidence interval were also considered. These 46 protein features specified 20 unique proteins. The remaining protein features represented multiple isoforms. Table 2 is a list of the identified protein features that displays the relative quantitative changes with their correlating probability statistics for each time point comparison. Twelve of 20 proteins were substantially changing among all burn time points within the 95th percentile (Student’s t test; p < 0.05; Table 3). In addition to these 12 proteins, 4 other proteins were also changed when Early was compared to Normal/Unwounded skin and a sole protein was changed when Middle was compared to Normal/Unwounded (p < 0.05). All 20 proteins were changed when Late wound samples were compared against levels in normal/unwounded skin (p < 0.05). Though each individual protein spot is analyzed independent of the other spots on each gel and p-values reflect the probability that the individual observed change occurred randomly, without confirming the statistics with a larger sample set, we present these finding as a trend towards significance.
Table 2.
Listing of identified proteins with their abundance changes from Normal to Late and the associated statistics
| Protein Identifications | Normal vs Late | ||
|---|---|---|---|
| Av. Ratio | T-test | ANOVA | |
| P08865 40S ribosomal protein SA | 3.08 | 0.013 | 0.0055 |
| serum albumin [fragment] | −3.36 | 0.03 | 0.077 |
| P60842 Eukaryotic initiation factor 4A–I | 1.67 | 0.0091 | 0.13 |
| * P02675 Fibrinogen beta | 16.01 | 0.00022 | 0.00012 |
| * P02679 Fibrinogen gamma | 9.67 | 0.00071 | 4.60E-05 |
| P06396 Gelsolin | −2.14 | 0.013 | 0.072 |
| P00738 Haptoglobin | 4.76 | 0.0042 | 0.00065 |
| P08238 Heat shock protein 90-beta | 7.6 | 0.0044 | 0.0023 |
| * P04264 Keratin 1 | −4.13 | 0.00082 | 0.0036 |
| * P13647 Keratin 5 | 10.11 | 0.014 | 0.025 |
| * P48668 Keratin 6 | 13.71 | 0.0036 | 0.0027 |
| * P08779 Keratin 16 | 12.12 | 5.20E-05 | 0.014 |
| * Q04695 Keratin 17 | 27.31 | 1.30E-05 | 0.0014 |
| * P07195 Lactate dehydrogenase B | 4.09 | 0.00049 | 0.00092 |
| P30740 Leukocyte elastase inhibitor | 8.08 | 7.80E-05 | 0.0065 |
| P46940 Ras GTPase-activating-like protein IQGAP1 | 1.9 | 0.002 | 0.033 |
| P29508 Squamous cell carcinoma antigen 1 | 23.83 | 0.00064 | 0.00014 |
| P48594 Squamous cell carcinoma antigen 2 | 20.73 | 0.0021 | 0.00035 |
| P13693 Translationally controlled tumor protein | 3.02 | 0.002 | 0.013 |
| * P08670 Vimentin | −3.32 | 0.043 | 0.019 |
Proteins previously identified with burn wounds in the literature.
Acute Stress Proteins and Metabolic Responses to Injury
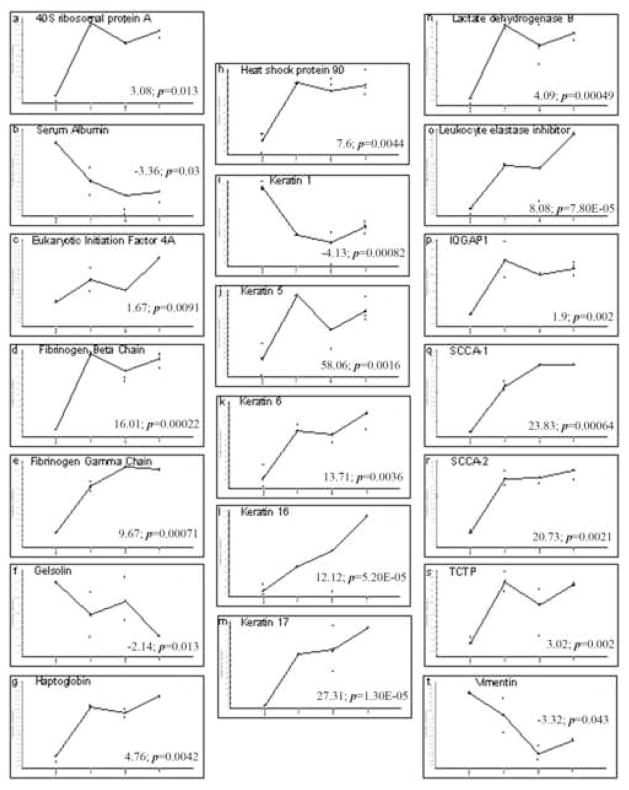
Five proteins were identified in this category. Heat Shock Protein (HSP90) was upregulated 6–7 fold in all burn periods (ANOVA p = 0.0023; Table 2 and Figure 2h). Lactate dehydrogenase B was identified and showed a 4-fold increase in expression (p < 0.001; Table 2 and Figure 2n). Five isoforms each were identified as fibrinogen beta and gamma chains. All isoforms of both displayed increases across the burn time points (Table 2 and Figures 2d and 2e). Serum albumin was also identified in our samples and showed a 3 fold decrease at the late time point (p = 0.03; Table 2 and Figure 2b).
Figure 2.
Graphical representation of the abundance changes for each protein. Representative graphs were selected for a single isoform for those proteins which matched to more than one protein spot. Samples are graphed from left to right in the following order, Normal/Unwounded, Early, Middle, and Late. All graphs are expressed in log scale. The fold change from Normal/Unwounded (far left of graph) to Late (far right of graph) and the associated student’s t-test have been included as an indication of scale.
Proteins Associated with the Inflammatory Response
Five proteins were also identified in this category. Haptoglobins were upregulated in isolates from burn wounds in our study (Table 2 and Figure 2g). The quantitative rise for five isoforms of this molecule hovered between 2–6 fold in wounds from our seriously burned cohort. Three different serine proteinase inhibitors (SLPI, SCCA1, and SCCA2) were identified. This is the first identification of leukocyte elastase inhibitor (SLPI) in human wounds. In our analysis, SLPI was elevated in all 3 burn timepoints (p = 0.0065; Table 2) but showed an 8-fold increase in the later burn period compared to the constitutive level observed in normal/unwounded skin (p < 0.0001; Table 2 and Figure 2o). Two isoforms each for the remaining two serpins, squamous cell carcinoma antigens 1 and 2 (SCCA1 and 2), are increased throughout the burn periods and both show greatest increases in the later burn period (Table 2 and Figures 2q and 2r). Translationally controlled tumor protein (TCTP) was also identified and was equally upregulated across all the burn periods in our study (as much as 3-fold, ANOVA p = 0.013; Table 2 and Figure 2s). Although, the data for the Middle timepoint are questionable due to the high variance we discovered through Principal Component Analysis.
Proteins Associated with Migrating and Proliferating Activities
Four proteins were identified in this category. Gelsolin in our study showed a slight decrease throughout all 3 burn periods (Figure 2f); however, this mild decrease was strongest in the later time point (p = 0.013; Table 2). Another cancer associated protein, 40S ribosomal protein SA (34/67-kDa laminin receptor) was detected in burn wounds, and was increased across all time points as compared to normal/unwounded (as much as 3-fold, ANOVA p = 0.0055; Table 2, Figure 2a). Eukaryotic initiation factor (eIF4A) was identified but was not substantially increased in the first two burn periods and only marginally upregulated in the later burn samples (1.67 fold, p = 0.0091; Table 2, Figure 2c). Lastly, GTPase activating protein (IQGAP1) was uniformly increased throughout all periods of the burn (p = 0.033; Table 2 and Figure 2p).
Cytoskeletal Proteins and Maintenance of Cellular Integrity
Six proteins were identified in this category and confirmed that our samples were isolated from the healing compartment of the burn. As expected, four isoforms for keratin 1 showed marked declines throughout postburn days 1–10 (Table 2 and Figure 2i). Two isoforms for keratin 5 were elevated throughout the time points (Table 2 and Figure 2j). Keratins 6, 16, and 17 also increased over time after injury. Nine isoforms of keratin 6 were detected, all of which increased throughout our time points (Table 2 and Figure 2k). Keratin 16 showed a 2 fold increase in the earliest wounds (p = 0.027) that eventually reached a 12 fold increase in the later burn period (p = 0.0001; Table 2 and Figure 2l). Keratin 17 showed a 10 fold increase in the earliest wounds (p = 0.0096) to a 27 fold increase in the later burn period (p < 0.0001; Table 2 and Figure 2m). Vimentin was also observed in our burn samples and showed a 2–5 fold decrease during the three temporal groupings (p = 0.019; Table 2, Figure 2t).
Discussion
The present manuscript provides evidence that DIGE/MS technology can be a useful discovery tool to elucidate molecular changes that occur after burn injury. Our evaluation uncovered forty-six statistically significant protein alterations. The principal component analysis methodology clustered the protein array and defined distinct temporal periods that develop during days 0–10 in the human burn wound. Although perturbations that occur after burn injury are exceedingly complex, the present study offers a new divide-and-conquer screening approach that should eventually supply the necessary precision to define the temporal sequence of a burn response. Of the twenty differentially expressed proteins, 9 were predicted from the literature but 10 were previously unassociated with either burn injury or wound healing. The implications for our eclectic findings using the DIGE/MS discovery tool are multi-focal and are discussed below in relationship to various known functional pathways evoked after injury or during wound repair.
Acute Stress Proteins and Metabolic Responses to Injury
Our data uncovered evidence of early response proteins. One example was heat shock protein 90 (HSP90), a molecular chaperone essential to the quality control pathway. This protein has previously been associated with thermal injury and hyperthermia(14, 15), therefore its elevation was expected. Another expected molecule was lactate dehydrogenase B. Its increased expression confirmed that the samples were harvested from the site of injury as it has long been considered a non-specific indicator of metabolic deficiency under ischemic circumstances that lead to tissue damage (16). Both fibrinogen components are major players in the wound healing process. Endothelial cells and fibroblasts are known to bind with fibrinogen in order to invade the fibrin clot formed at injury sites (17, 18). It has also been suggested that fibrinogen aids in the sloughing of wound eschar because keratinocytes lack the receptors necessary for binding to fibrinogen and therefore, they migrate beneath the fibrin clot (17).
Proteins Associated with the Inflammatory Response
The inflammatory process is quite pronounced in burn wounds so it was not surprising to find a large assortment of proteins fulfilling these functions. Haptoglobin, a molecule known to bind free hemoglobin thereby limiting oxidative damage to tissues, was identified for the first time in human burn wounds. Nevertheless, there are multiple clues that explain its possible mechanism of action. Haptoglobin reportedly serves as a cell migration factor for endothelial cells in vascular repair, which is certainly a component of wound repair (19), and tissue expression of haptoglobin from local fibroblasts can be elevated in the presence of LPS (19), a bacterial product which one might expect in these wounds.
The involvement of serine proteinases (elastase) in burn wounds and fluid was originally noted in 1987 and was later confirmed by the detection of proteinase inhibitors in burn fluid (20, 21). SLPI is a potent inhibitor of serine proteases such as neutrophil elastase, cathepsin G, and mast cell chymase and is one of many innate immunity-associated proteins (22, 23). Sources of SLPI in the wound environment are plentiful, and include neutrophils (22), macrophages (24, 25), and keratinocytes (26–28). Emerging roles for SLPI are equally numerous and it has been suggested that SLPI could be used as a marker to monitor the progress of infection (22). Mice with a null phenotype for SLPI show impaired cutaneous wound healing with increased inflammation and elastase activity (29). The later increase shown in our study correlates well with murine studies that have shown that macrophages secrete an increased amount of SLPI when encountering apoptotic cells which may help to attenuate inflammation during the clearance of these cells (25).
Molecules Previously Associated with Tumor Development
The presence of several cancer related proteins was unexpected. Among these are SCCA’s. Although SCCA’s have been characterized as serological markers of advanced squamous cell carcinomas, they apparently have numerous potential functions within the burn wound. Wound beds have enormous local concentration of elastases and cathepsins G molecules due to leukocyte activation. SCCA1 is an inhibitor of papain-like cysteine proteinases such as cathepsins I, S, and K (30), and SCCA2 inhibits the chymotrypsin-like proteinases such as cathepsin G and mast cell chymase (31). These molecules have been validated as biomarkers that reflect the clinical severity of the Th2 response in atopic dermatitis (32), and their serum levels correlate well with lactate dehydrogenase, another protein identified as upregulated in this initial report. In normal skin, these proteins are distributed in the outer root sheath of hair follicles (30) and their secretion from cultured keratinocytes has been documented (33). SCCA1 acts to protect UV-exposed (stressed) keratinocytes from apoptosis (34). SCCA-1 and 2 have been associated with several different forms of cancer, but never directly with the rare squamous cell carcinomas development in burn tissue (35, 36). Thus this molecule is a newly discovered candidate protein in the burn wound.
Translationally controlled tumor protein (TCTP) has previously been characterized as a secreted protein in epidermal keratinocyte cultures (33). Growth signals, cytokine expression, and stress conditions have been shown to influence TCTP synthesis (37), so its up-regulation is not a surprise, though its potential roles in burn wound healing are not fully characterized.
Proteins Associated with Migrating and Proliferating Activities
We were reassured that our protein analysis was entirely valid when a diversity of migratory and proliferating associated proteins were detected. Gelsolin is a cytoplasmic, calcium regulated, actin modulating protein in keratinocytes and fibroblasts that binds to actin filament ends and can mediate cell migration (38, 39). Work with gelsolin null mice (40) established the requirement of gelsolin for rapid motile cell responses to stress, such as hemostasis, inflammation and wound healing. Gelsolin is well characterized as a pivotal molecule to EGF-stimulated fibroblast migration (38, 39). The wound periods in our study were relatively early phases of a lengthy repair process greatly preceding the scar formation period, and therefore may indicate either a deficiency in the wound repair process or simply that scar formation has not yet commenced.
The limited evidence that 40S ribosomal protein SA (34/67-kDa laminin receptor) is present in the skin indicates its prevalence in areas of keratinocyte proliferation (41), so the mild increase in the wound bed was expected. However, its increased expression has been more commonly associated with metastatic tumors (42). We speculate that many molecules associated with neoplastic growth are also non-specific players in the transient type of benign proliferation seen in the wound healing response.
GTPase activating protein (IQGAP1) interacts with components of the cytoskeleton, cell-adhesion, and signaling molecules as it regulates cell morphology. Two specific functions make it likely that this protein is pivotal to wound repair. This negative regulator of cell-cell adhesion allows epidermal and endothelial cells at the periphery of the burn to detach from their adjacent neighbors and migrate into the wound defect (43). Secondly it is known to interact with Rac1, cdc42 and actin filaments during polarization and migration of fibroblasts and keratinocytes. Such activities are plentiful in the burn wound and confirms the validity of our techniques to characterize the burn wound(44).
Cytoskeletal Proteins and Maintenance of Cellular Integrity
Previously characterized structural proteins (keratins 6, 16, 17) were detected and confirmed the validity of the assays (34, 47, 48, 49, 50). Vimentin intermediate filaments are important for stabilizing the architecture of the cytoplasm in all mesenchymal cells and a decline was predicted from our earlier immunohistochemical staining after burn injury where we used this protein as a marker to delineate the downward progression in tissue damage following burn injury (2, 45).
Additional Observations
Our initial proteomic survey of wound healing in humans was notable from the standpoint of those proteins that were not detected. For example, none of the 46 highly expressed protein spots were growth factor receptors or other transmembrane spanning larger protein molecules. This was a predictable finding considering their higher molecular weights and solubility coefficients that most likely excluded them from the initial isolates. Other proteins such as those with low copy number, highly acidic/basic properties were likewise excluded from the analysis, as highly hydrophobic molecules or those that are less than 10kDa or approaching 200kDa are not easily captured by the techniques used in this manuscript. Future studies with complementary techniques designed to detect both larger (ex. transmembrane receptors) and smaller (ex. cytokines) molecules will be necessary to obtain a comprehensive understanding of signaling networks, extracellular matrix molecules, and post-translational changes such as glycosylation or phosphorylation that are also evoked following injury and during the healing response.
In summary, this preliminary study assessed the ability of DIGE/MS analysis to distinguish among post burn day timepoints based on protein expression changes isolated from burn samples and resolved on 2D gels. Our future goals are to expand our patient sampling, use complementary techniques to evaluate proteins not detectable by DIGE methods, and confirm our findings with immunohistochemistry. The present study is based on the premise that identification of the protein patterns that define the critical events after burn injury and the ensuring wound repair may disclose potential therapeutic targets.
Acknowledgments
The authors thank Drs. Jeffrey Guy, Adam Ellis, and Blair Summitt for access to patients in Vanderbilt’s Burn Center. We express appreciation to Joseph A. Greco, MD and Nancy Cardwell for assistance in sample collection. We also thank Corbin W. Whitwell for technical support for DIGE/MS and the Vanderbilt Academic Venture Capitol fund for support for proteomics through the Proteomics Laboratory of the Vanderbilt Ingram Cancer Center (5 P30 CA 068485-10). Financial support was provided by National Institute of Health grant GM 40437 (ACP, LBN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gibran NS, Heimbach DM. Current status of burn wound pathophysiology. Clin Plast Surg. 2000;27:11–22. [PubMed] [Google Scholar]
- 2.Nanney LB, Wenczak BA, Lynch JB. Progressive burn injury documented with vimentin immunostaining. J Burn Care Rehabil. 1996;17:191–198. doi: 10.1097/00004630-199605000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Herndon DN. Total Burn Care. London: W.B. Saunders; 2002. [Google Scholar]
- 4.Harrison CA, Gossiel F, Bullock AJ, Sun T, Blumsohn A, Mac Neil S. Investigation of keratinocyte regulation of collagen I synthesis by dermal fibroblasts in a simple in vitro model. Br J Dermatol. 2006;154:401–410. doi: 10.1111/j.1365-2133.2005.07022.x. [DOI] [PubMed] [Google Scholar]
- 5.Lofts JA. Cost analysis of a major burn. N Z Med J. 1991;104:488–490. [PubMed] [Google Scholar]
- 6.Akita S, Akino K, Imaizumi T, Hirano A. A basic fibroblast growth factor improved the quality of skin grafting in burn patients. Burns. 2005;31:855–858. doi: 10.1016/j.burns.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Mann R, Heimbach D. Prognosis and treatment of burns. West J Med. 1996;165:215–220. [PMC free article] [PubMed] [Google Scholar]
- 8.Brigham PA, McLoughlin E. Burn incidence and medical care use in the United States: estimates, trends, and data sources. J Burn Care Rehabil. 1996;17:95–107. doi: 10.1097/00004630-199603000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Ibuki Y, Naitou H, Ohashi N, Goto R. Proteome analysis of UV-B-induced anti-apoptotic regulatory factors. Photochem Photobiol. 2005;81:823–829. doi: 10.1562/2004-09-01-RA-297. [DOI] [PubMed] [Google Scholar]
- 10.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 11.Lilley KS, Friedman DB. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev Proteomics. 2004;1:401–409. doi: 10.1586/14789450.1.4.401. [DOI] [PubMed] [Google Scholar]
- 12.Friedman DB, Wang SE, Whitwell CW, Caprioli RM, Arteaga CL. Multi-variable Difference Gel Electrophoresis and Mass Spectrometry: A Case Study on TGF-beta and ErbB2 signaling. Mol Cell Proteomics. 2006 doi: 10.1074/mcp.D600001-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Ogura H, Hashiguchi N, Tanaka H, Koh T, Noborio M, Nakamori Y, Nishino M, Kuwagata Y, Shimazu T, Sugimoto H. Long-term enhanced expression of heat shock proteins and decelerated apoptosis in polymorphonuclear leukocytes from major burn patients. J Burn Care Rehabil. 2002;23:103–109. doi: 10.1097/00004630-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Diao L, Li C, Sheng Z. The significance of the expression of the HSP70 and HSP90 in the intestinal mucosa in scalded rats during early postburn stage. Zhonghua Shao Shang Za Zhi. 2000;16:279–282. [PubMed] [Google Scholar]
- 16.Kabasakal L, Sener G, Cetinel S, Contuk G, Gedik N, Yegen BC. Burn-induced oxidative injury of the gut is ameliorated by the leukotriene receptor blocker montelukast. Prostaglandins Leukot Essent Fatty Acids. 2005;72:431–440. doi: 10.1016/j.plefa.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Kubo M, Van de Water L, Plantefaber LC, Mosesson MW, Simon M, Tonnesen MG, Taichman L, Clark RA. Fibrinogen and fibrin are anti-adhesive for keratinocytes: a mechanism for fibrin eschar slough during wound repair. J Invest Dermatol. 2001;117:1369–1381. doi: 10.1046/j.0022-202x.2001.01551.x. [DOI] [PubMed] [Google Scholar]
- 18.Mosesson MW, Siebenlist KR, Meh DA. The structure and biological features of fibrinogen and fibrin. Ann N Y Acad Sci. 2001;936:11–30. doi: 10.1111/j.1749-6632.2001.tb03491.x. [DOI] [PubMed] [Google Scholar]
- 19.de Kleijn DP, Smeets MB, Kemmeren PP, Lim SK, Van Middelaar BJ, Velema E, Schoneveld A, Pasterkamp G, Borst C. Acute-phase protein haptoglobin is a cell migration factor involved in arterial restructuring. Faseb J. 2002;16:1123–1125. doi: 10.1096/fj.02-0019fje. [DOI] [PubMed] [Google Scholar]
- 20.Grinnell F, Zhu M. Identification of neutrophil elastase as the proteinase in burn wound fluid responsible for degradation of fibronectin. J Invest Dermatol. 1994;103:155–161. doi: 10.1111/1523-1747.ep12392625. [DOI] [PubMed] [Google Scholar]
- 21.Faymonville ME, Micheels J, Bodson L, Jacquemin D, Lamy M, Adam J, Duchateau J. Biochemical investigations after burning injury: complement system, protease-antiprotease balance and acute-phase reactants. Burns Incl Therm Inj. 1987;13:26–33. doi: 10.1016/0305-4179(87)90252-x. [DOI] [PubMed] [Google Scholar]
- 22.Doumas S, Kolokotronis A, Stefanopoulos P. Anti-inflammatory and antimicrobial roles of secretory leukocyte protease inhibitor. Infect Immun. 2005;73:1271–1274. doi: 10.1128/IAI.73.3.1271-1274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Nathan C, Jin W, Sim D, Ashcroft GS, Wahl SM, Lacomis L, Erdjument-Bromage H, Tempst P, Wright CD, Ding A. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111:867–878. doi: 10.1016/s0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 24.Thuraisingam T, Sam H, Moisan J, Zhang Y, Ding A, Radzioch D. Delayed cutaneous wound healing in mice lacking solute carrier 11a1 (formerly Nramp1): correlation with decreased expression of secretory leukocyte protease inhibitor. J Invest Dermatol. 2006;126:890–901. doi: 10.1038/sj.jid.5700182. [DOI] [PubMed] [Google Scholar]
- 25.Odaka C, Mizuochi T, Yang J, Ding A. Murine macrophages produce secretory leukocyte protease inhibitor during clearance of apoptotic cells: implications for resolution of the inflammatory response. J Immunol. 2003;171:1507–1514. doi: 10.4049/jimmunol.171.3.1507. [DOI] [PubMed] [Google Scholar]
- 26.Sorensen OE, Cowland JB, Theilgaard-Monch K, Liu L, Ganz T, Borregaard N. Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J Immunol. 2003;170:5583–5589. doi: 10.4049/jimmunol.170.11.5583. [DOI] [PubMed] [Google Scholar]
- 27.Lai JY, Borson ND, Strausbauch MA, Pittelkow MR. Mitosis increases levels of secretory leukocyte protease inhibitor in keratinocytes. Biochem Biophys Res Commun. 2004;316:407–410. doi: 10.1016/j.bbrc.2004.02.065. [DOI] [PubMed] [Google Scholar]
- 28.Wingens M, van Bergen BH, Hiemstra PS, Meis JF, van Vlijmen-Willems IM, Zeeuwen PL, Mulder J, Kramps HA, van Ruissen F, Schalkwijk J. Induction of SLPI (ALP/HUSI-I) in epidermal keratinocytes. J Invest Dermatol. 1998;111:996–1002. doi: 10.1046/j.1523-1747.1998.00425.x. [DOI] [PubMed] [Google Scholar]
- 29.Ashcroft GS, Lei K, Jin W, Longenecker G, Kulkarni AB, Greenwell-Wild T, Hale-Donze H, McGrady G, Song XY, Wahl SM. Secretory leukocyte protease inhibitor mediates non-redundant functions necessary for normal wound healing. Nat Med. 2000;6:1147–1153. doi: 10.1038/80489. [DOI] [PubMed] [Google Scholar]
- 30.Cataltepe S, Gornstein ER, Schick C, Kamachi Y, Chatson K, Fries J, Silverman GA, Upton MP. Co-expression of the squamous cell carcinoma antigens 1 and 2 in normal adult human tissues and squamous cell carcinomas. J Histochem Cytochem. 2000;48:113–122. doi: 10.1177/002215540004800112. [DOI] [PubMed] [Google Scholar]
- 31.Schick C, Kamachi Y, Bartuski AJ, Cataltepe S, Schechter NM, Pemberton PA, Silverman GA. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J Biol Chem. 1997;272:1849–1855. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- 32.Mitsuishi K, Nakamura T, Sakata Y, Yuyama N, Arima K, Sugita Y, Suto H, Izuhara K, Ogawa H. The squamous cell carcinoma antigens as relevant biomarkers of atopic dermatitis. Clin Exp Allergy. 2005;35:1327–1333. doi: 10.1111/j.1365-2222.2005.02353.x. [DOI] [PubMed] [Google Scholar]
- 33.Katz AB, Taichman LB. A partial catalog of proteins secreted by epidermal keratinocytes in culture. J Invest Dermatol. 1999;112:818–821. doi: 10.1046/j.1523-1747.1999.00572.x. [DOI] [PubMed] [Google Scholar]
- 34.Katagiri C, Nakanishi J, Kadoya K, Hibino T. Serpin squamous cell carcinoma antigen inhibits UV-induced apoptosis via suppression of c-JUN NH2-terminal kinase. J Cell Biol. 2006;172:983–990. doi: 10.1083/jcb.200508064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akiyama M, Inamoto N, Nakamura K. Malignant melanoma and squamous cell carcinoma forming one tumour on a burn scar. Dermatology. 1997;194:157–161. doi: 10.1159/000246086. [DOI] [PubMed] [Google Scholar]
- 36.Lee SH, Shin MS, Kim HS, Park WS, Kim SY, Jang JJ, Rhim KJ, Jang J, Lee HK, Park JY, Oh RR, Han SY, Lee JH, Lee JY, Yoo NJ. Somatic mutations of Fas (Apo-1/CD95) gene in cutaneous squamous cell carcinoma arising from a burn scar. J Invest Dermatol. 2000;114:122–126. doi: 10.1046/j.1523-1747.2000.00819.x. [DOI] [PubMed] [Google Scholar]
- 37.Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP) Int J Biochem Cell Biol. 2004;36:379–385. doi: 10.1016/s1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 38.Wells A, Ware MF, Allen FD, Lauffenburger DA. Shaping up for shipping out: PLCgamma signaling of morphology changes in EGF-stimulated fibroblast migration. Cell Motil Cytoskeleton. 1999;44:227–233. doi: 10.1002/(SICI)1097-0169(199912)44:4<227::AID-CM1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 39.Xie H, Pallero MA, Gupta K, Chang P, Ware MF, Witke W, Kwiatkowski DJ, Lauffenburger DA, Murphy-Ullrich JE, Wells A. EGF receptor regulation of cell motility: EGF induces disassembly of focal adhesions independently of the motility-associated PLCgamma signaling pathway. J Cell Sci. 1998;111 (Pt 5):615–624. doi: 10.1242/jcs.111.5.615. [DOI] [PubMed] [Google Scholar]
- 40.Witke W, Sharpe AH, Hartwig JH, Azuma T, Stossel TP, Kwiatkowski DJ. Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin. Cell. 1995;81:41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- 41.Cavalieri S, Rotoli M, Feliciani C, Amerio P. Expression of the high-affinity laminin receptor (67 kDa) in normal human skin and appendages. Int J Immunopathol Pharmacol. 2005;18:223–231. doi: 10.1177/039463200501800205. [DOI] [PubMed] [Google Scholar]
- 42.Zhou L, Xie M, Zhou JQ, Tao L. 67-kDa laminin receptor in human laryngeal squamous cell carcinoma. Laryngoscope. 2006;116:28–32. doi: 10.1097/01.mlg.0000184505.05662.b9. [DOI] [PubMed] [Google Scholar]
- 43.Sugimoto N, Imoto I, Fukuda Y, Kurihara N, Kuroda S, Tanigami A, Kaibuchi K, Kamiyama R, Inazawa J. IQGAP1, a negative regulator of cell-cell adhesion, is upregulated by gene amplification at 15q26 in gastric cancer cell lines HSC39 and 40A. J Hum Genet. 2001;46:21–25. doi: 10.1007/s100380170119. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 45.Ikegawa S, Saida T, Takizawa Y, Tokuda Y, Ito T, Fujioka F, Sakaki T, Uchida N, Arase S, Takeda K. Vimentin-positive squamous cell carcinoma arising in a burn scar. A highly malignant neoplasm composed of acantholytic round keratinocytes. Arch Dermatol. 1989;125:1672–1676. [PubMed] [Google Scholar]