Fig. 9.
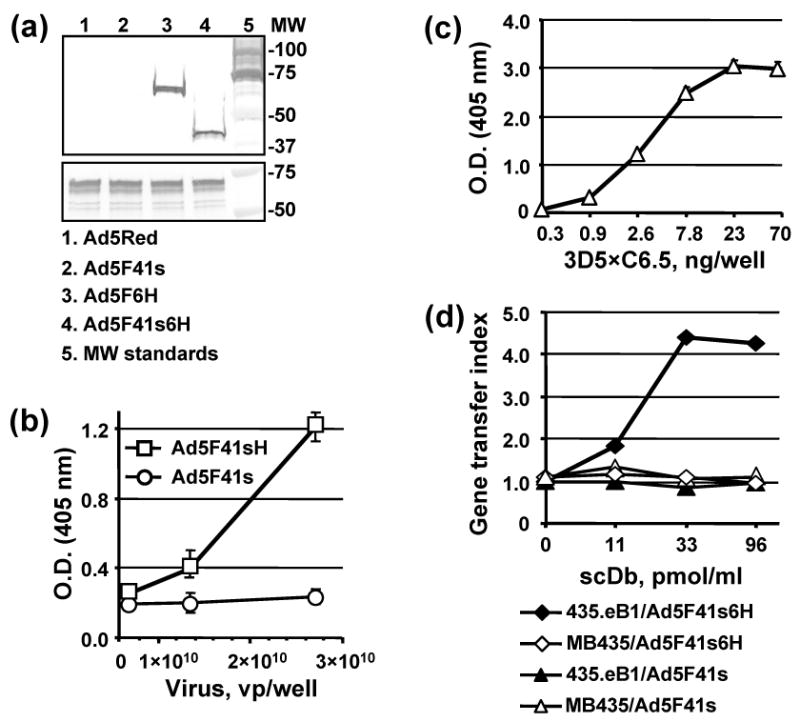
Characterization of 3D5×C6.5 scDb binding specificity. (a) Electrophoretically resolved viral proteins were transferred to a PVDF membrane and probed with 3D5×C6.5 scDb, diluted to a concentration of 1 μg/ml, or penton-base specific rabbit sera to detect the presence of His6-tagged fiber proteins (upper panel) or penton base (lower panel) respectively. The numbers on the right indicate molecular masses of protein standards (lane 5) in kDa. (b) Ad5F41s6H or Ad5F41s virions immobilized on an ELISA plate at the indicated concentrations were incubated with 100 ng/ml of 3D5×C6.5 scDb. Bound scDb protein was detected with an anti-HA mAb conjugated with AP. (c) 3D5×C6.5 scDb was diluted to the indicated concentrations, adsorbed on an ELISA plate, and incubated with 50 μl aliquots of SKBR-3 cell lysate. Bound c-erbB2 oncoprotein was detected with mAb cocktail against the cytoplasmic c-erbB2 domain followed by AP-conjugated goat anti-mouse secondary antibody, and the plate was read at 405 nm. (d) Samples of A5F41s6H and A5F41s vectors were preincubated with the indicated dilutions of 3D5×C6.5 scDb for 30 min and then used to infect 435.eB1 or MDA-MB-435 cells at a dose of 100 vp/cell. The level of DsRed2 reporter gene expression was assessed two days postinfection by determining the RFU values using 560 nm emission and 620 nm excitation filters. The ratios of RFU detected in the cells infected in the presence of scDb adapter to RFU detected in cells infected with virus alone were calculated and are presented as gene transfer indices.
