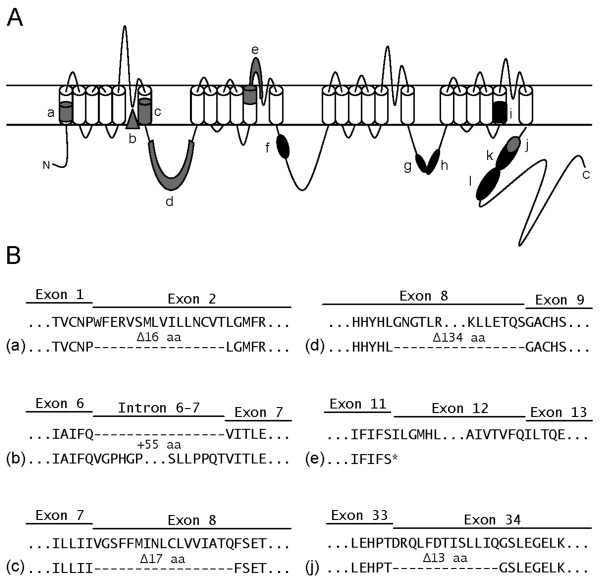
Figure 3.
Alternative splicing architecture of the mouse Cacna1g gene. (A) Representation of the Cav3.1/α1G T-type calcium channel structure and the localization of the 12 alternatively spliced regions: a) Δ5' E2; b) +I6-7; c) Δ5' E8; d) Δ3' E8; e) ΔE12; f) ΔE14; g) Δ3' E25; h) ΔE26; i) Δ5' E31; j) Δ5' E34; k) ΔE34; and l) ΔE35. Regions in black correspond to the previously identified alternative splice variants [9,12] and segments in grey denote the newly-characterized splice variants detected in this study. (B) Predicted protein sequences arising from the a), b), c), d), e), and j) alternative splicing events compared to full length Cav3.1/α1G protein, dashes indicate in-frame amino acid exclusion and the asterisk signifies a nonsense codon truncation.