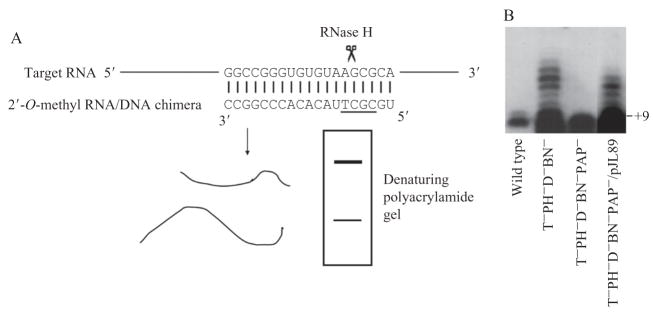
Figure 2.2.
Analysis of RNA termini by site-directed RNase H cleavage. (A) Illustration of the method. A chimeric oligonucleotide containing a stretch of DNA (underlined sequence) is used to direct RNase H cleavage at a position close to the 3′ end of 23S rRNA. (B) The 3′ ends of precursor to 23S RNA revealed by site-directed RNase H cleavage (adapted from Li et al., [1999] with permission). The products were separated in a denaturing polyacrylamide gel and were detected by Northern blotting with a probe specific for the 3′ terminal sequence of 23S RNA. A small amount of precursor with eight extra nucleotides at the 3′ end is present in 23S RNA from wild-type cells. In a mutant strain deficient in the exoribonucleases, RNase T, PH, D, and BN (T−PH−D−BN−), a precursor containing 9 or more 3′ extra residues was found. These longer products are formed by polyadenylation. Removal of poly(A) polymerase from the multi-RNase–deficient strain (T−PH−D−BN−PAP−) restored the precursor to +8nt size. Introduction of the plasmid pJL89 harboring the poly(A) polymerase gene into the PAP− background (T−PH−D−BN−PAP−/pJL89) resulted in formation of the polyadenylated species.