Fig. 8.
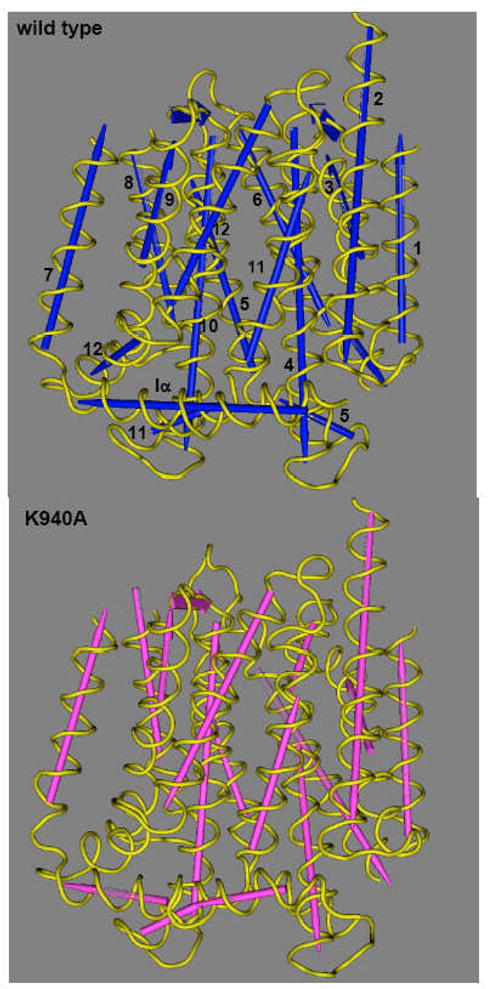
Structure of transmembrane domains of AcrB proton-relay mutants. The figure was drawn by using the Cn3D program (National Center for Biotechnology Information) after alignment of the three-dimensional structures of the wild type (PDB file 1IWG) and the Lys940Ala mutant (this study) through the Vector Alignment Search Tool, available at the National Center for Biotechnology Information website. Note that the assignment of the secondary structure was done in a uniform manner by the Vector Alignment Search Tool program. The TM helices are numbered in the wild-type structure. Although not shown here, other mutants of the network residues (Asp407Ala, Asp408Ala, Thr978Ala) show very similar conformational alterations. From [53].
