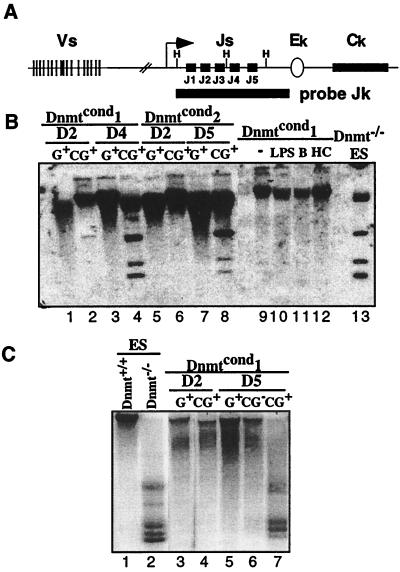
Figure 3.
Loss of Dnmt1 results in the loss of genomic methylation by day 4 postinfection. (A) Schematic diagram of the murine kappa locus. The map shows the V and J gene segments, constant region (Ck) and the intronic enhancer (Ek). The methylation-sensitive restrictions sites, HhaI (H), are shown as well as the probe used for analysis (Jk). The diagram is not to scale. (B) Southern blot analysis of the methylation status of the kappa locus in Abelson lines under different conditions. The DNA was digested with HhaI and probed with a Jk probe that spans the J region of the kappa locus. Lanes 1–8 are derived from Gfp+ sorted Abelson lines infected with the MiGfp (G) or MCiGfp (CG) at the time points indicated postinfection. Lanes 9–12 are generated from unsorted bulk populations of cells. Lane 9, untreated; lane 10, treated with LPS; lane 11, treated with butyrate; lane 12 treated with HC toxin. Lane 13 is derived from untreated Dnmt−/− ES cells. (C) Southern blot analysis of the methylation status of IAPs in Abelson lines. The DNA was digested with HpaII and probed with a fragment of a known, endogenous IAP. Lanes 1 and 2 are controls, derived from ES cells that are wild type or mutant for the Dnmt1 locus, respectively. Lanes 3–7 are derived from Gfp+ or Gfp− sorted Dnmtcond1 Abelson line infected with either MiGfp (G) or MCiGfp (CG) at the time points indicated. The unmethylated fragments are shown on the right.