Melanopsin (Opn4) contributes to the entrainment of circadian rhythms in mammals. In rats, retinal ganglion cells expressing Opn4 sense light and regulate circadian entrainment (Hattar et al., 2002) by relaying light information to the SCN via release of neuropeptides, including glutamate and pituitary adenylate cyclase-activating peptide (PACAP) (Hannibal, 2006). Opn4 mRNA has also been found in horizontal and/or ganglion cells in teleost fish, including zebrafish, roach, and Atlantic cod, but in these species its function is unknown.
Vertebrate ancient opsin (VA opsin), first found in Atlantic salmon (Soni and Foster, 1997) and subsequently in zebrafish, carp, smelt, and roach, may also contribute to circadian entrainment in teleosts. In roach, one type of horizontal cell is intrinsically photosensitive and depolarizes maximally in response to light of wavelengths matching VA opsin sensitivity (Jenkins et al., 2003), suggesting that VA opsin can provide photic information to the brain.
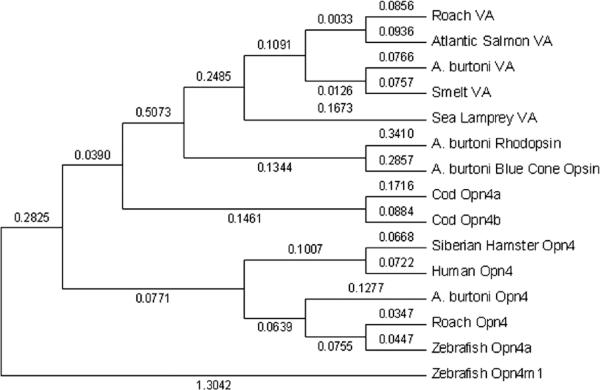
We cloned and sequenced Opn4, VA opsin, and PACAP from an African cichlid fish, Astatotilapia burtoni (Fig. 1). The A. burtoni Opn4 amino acid transmembrane region resembles that of zebrafish Opn4a (77.4% identity) more than zebrafish Opn4m1 (67.1%). The 391-amino acid A. burtoni VA opsin is homologous to the VA-L isoform found in smelt (Minamoto and Shimizu, 2002). A.burtoni PACAP cDNA differs at only 2 base pairs (of 599) from tilapia (Oreochromis mossambicus) PACAP (AY830121).
Figure 1.
Phylogenetic tree of vertebrate ancient (VA) opsin and Opn4 protein sequences, using zebrafish Opn4m1 as the outgroup. Tree generated using neighbor-joining methods based on 284 amino acids of transmembrane domains. Bootstrap values using 500 replicates are shown.
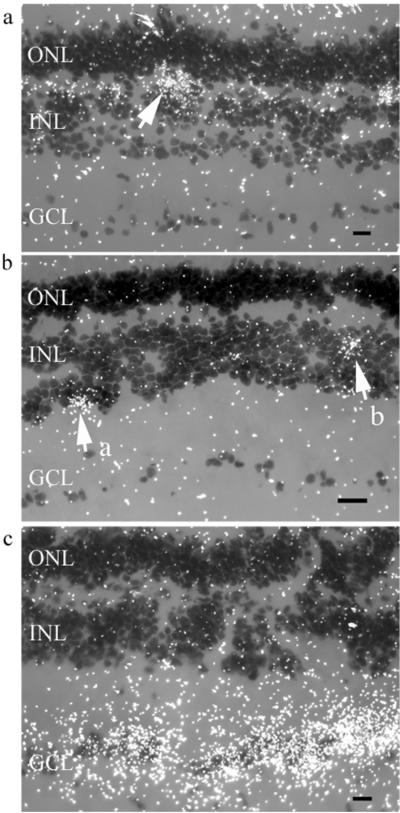
Based on cell morphology, Opn4 and VA were suggested to be coexpressed in roach (Jenkins et al., 2003) and Atlantic cod retinal neurons (Drivenes et al., 2003). We localized Opn4 and VA opsin in A. burtoni retinae using in situ hybridization and did not find coexpression. Instead, we show that A. burtoni Opn4 mRNA expression is restricted to cells with the characteristic morphology and position of horizontal cells (Fig. 2a). Labeled cells were rare and approximately uniformly distributed across the retina. Opn4 expression in A. burtoni retina differs from that in Atlantic cod retina, where 2 Opn4 splicing forms are found in amacrine and ganglion cells (Drivenes et al., 2003). In cypriniformes, zebrafish and roach Opn4 expression patterns resemble those found in A. burtoni, with Opn4 in subsets of horizontal cells (Bellingham et al., 2002; Jenkins et al., 2003).
Figure 2.
Photomicrographs of darkfield images from typical in situ hybridizations of sections of Astatotilapia burtoni retina showing high concentrations of silver grains (white dots) identifying probe location. Background label is scattered white dots. Scale bar represents 10 μm. INL, inner nuclear layer; ONL, outer nuclear layer; GCL, ganglion cell layer. (a) Opn4 in situ probe shows expression in the outer plexiform layer (OPL), between INL and ONL, localized to horizontal cells. White arrow indicates a typical cluster of staining surrounding an Opn4-containing horizontal cell. (b) Vertebrate ancient (VA) opsin in situ probe reveals VA opsin expression localized in 2 subsets of cells. Arrow "a" indicates vitreal INL amacrine cells. Arrow "b" indicates bipolar cells positioned between amacrine and horizontal cells. (c) Pituitary adenylate cyclase-activating peptide (PACAP) in situ probe shows intense staining in retinal ganglion cells.
We localized VA opsin expression in 2 subsets of inner nuclear layer (INL) cells (Fig. 2b): (1) amacrine cells in the vitreal INL (indicated by arrow "a" in Fig. 2b) and (2) cells in the INL positioned between the amacrine and horizontal cell layers, suggesting they are bipolar cells (indicated by arrow "b" in Fig. 2b). Approximately 10 times more putative amacrine than bipolar cells, and no other cell types, were labeled. Similarly,in Atlantic salmon, VA opsin was reported in putative amacrine and horizontal cells (Philp et al., 2000). In 2 cypriniformes, zebrafish (Kojima et al., 2000) and roach (Jenkins et al.,2003), expression of VA opsin was reported in horizontal cells, and in roach it was also reported in some ganglion cells. In smelt, VA opsin is found exclusively in the amacrine cell layer (Minamoto and Shimizu, 2002).
PACAP-labeled cells were found expressed abundantly exclusively in the retinal ganglion cell (RGC) layer (Fig. 2c). Since A. burtoni retina sends direct projections to the SCN (Fernald, 1982), Opn4, VA opsin, and PACAP could stimulate SCN activity.
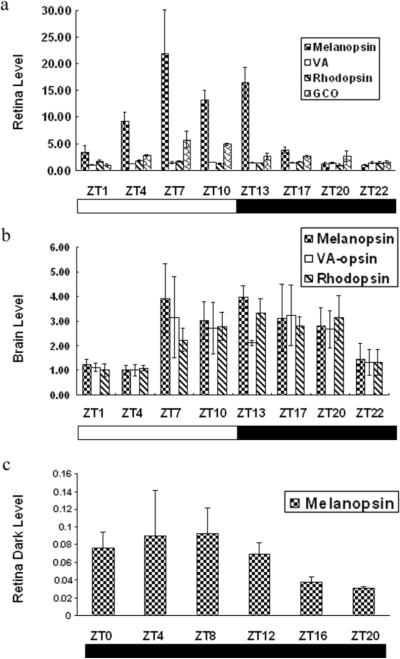
We measured the daily rhythm of Opn4, VA opsin, rhodopsin, and green cone opsin (GCO) mRNA levels in brain and retina using quantitative reverse transcription PCR (qRT-PCR). Opn4 mRNA levels in retina fluctuated significantly (p < 0.001), resembling the significant (p < 0.001) GCO rhythm (Fig. 3a). For both Opn4 and GCO, mRNA abundance was highest in the afternoon, between 7 and 10 h after onset of the light phase (i.e., ZT7 and ZT10). This Opn4 rhythm is in phase with that reported for mRNA expression in the rat retina (Park and Kang, 2006). VA opsin was slightly but not significantly elevated between ZT7 and ZT10. Rhodopsin mRNA was significantly elevated from ZT7 to ZT15 (p = 0.009), as previously reported (Korenbrot and Fernald, 1989). Levels of brain Opn4 and VA opsin mRNA did not fluctuate significantly (Fig. 3b).
Figure 3.
mRNA levels for Opn4, vertebrate ancient (VA) opsin, rhodopsin, and green cone opsin (GCO) measured as a function of time of day using quantitative reverse transcription PCR (qRT-PCR). Each bar in the histogram represents levels for 3 (a, b) or 5 (c) fish normalized to the geometric mean of β-tubulin and 18S rRNA concentration over 24 h, plotted as fold change with the lowest value taken as 1. Error bars represent 1 standard deviation. (a) Retina mRNA levels. (b) Brain mRNA levels. (c) Retinal mRNA levels of animals kept in constant darkness for 7 days.
To determine whether light is necessary for daily upregulation of Opn4 mRNA, A. burtoni were kept in constant darkness for 7 days and then retinal tissues were collected at 6 time points, every 4 hours around the clock. Retinal Opn4 mRNA levels in these animals did not differ between time points (Fig. 3c), showing that Opn4 mRNA changes are light dependent in A. burtoni.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Neurological Disorders and Stroke, NIH, NS 34950, the J. Javits Neuroscience Investigator Award to Russell D. Fernald. We thank Julie Desjardins, Wayne Korzan, Katharine Mach, Christopher Olin, Norman Ruby, Alex Vaughan, Martin Zatz, and 3 anonymous reviewers for useful comments on this manuscript.
APPENDIX MATERIALS AND METHODS
A. burtoni were kept in conditions similar to those in their native habitat, Lake Tanganyika (12-h light/12-h dark cycle; lights on at 0800 h PST; 27 °C; pH 8.0).
Animals were sacrificed by rapid cervical transaction, retinas removed, and mRNA extracted using Trizol (Invitrogen, Carlsbad, CA). cDNA for cloning was transcribed from total mRNA with a SMART RACE kit (Clontech, Mountain View, CA). Degenerate primers were designed to amplify Opn4 and VA opsin cDNA based on known Opn4 and VA opsin mRNA sequences. PACAP was cloned using nondegenerate primers based on the O. mossambicus PACAP cDNA sequence.
VA opsin species names and GenBank accession numbers: Plecoglossus altivelis: AB074483; Salmo salar: AF001499; Rutilus rutilus: AY116411; Petromyzon marinus: AAC41240 (originally P Opsin; Moutsaki et al., 2000). Opn4 accession numbers: Danio rerio opn4a: NM_178289; Danio rerio opn4m1: Q2KNE5; Rutilus rutilus: AY226847; Phodopus sungorus: AY726733; Homo sapiens: BC113558.
PCR products were sequenced (Sequetech, Mountain View, CA), and sequences were used to design primers for Rapid Amplification of cDNA Ends (RACE) PCR to clone the full-length cDNA (SMART RACE cDNA Amplification Kit, BD Biosciences, USA) from total retinal RNA. Reaction products were purified (QIAquick, Qiagen, Valencia, CA), subcloned (TOPO, Invitrogen), and sequenced (Sequetech).
Translated protein sequences were aligned and a 284-amino acid region containing the transmembrane domains was chosen for phylogenetic analysis. A tree for Opn4 and VA opsin was generated with neighbor-joining methods, and bootstrap values were calculated (MEGA 3.1; Kumar et al., 2004).
In situ hybridization followed standard procedures as modified by our laboratory, and counterstaining with Cresyl violet identified cell bodies. Sense versions of each probe were used to test the specificity, and signal above background was not present. Photomicrographs were captured digitally (SPOT camera system, Diagnostic Instruments, Sterling Heights, MI).
To assess variation in mRNA levels over 24 h, quantitative reverse transcription PCR (qRT-PCR) was performed (MyIQ Real-Time PCR Detection System, Bio-Rad Laboratories, Hercules, CA). During a light/dark cycle, brain and retinal tissues were extracted at 3-h intervals in a 24-h period from 3 animals killed and immediately dissected at each time point. Note: 1 brain from time point ZT13 was lost in processing. For the constant dark condition, 5 animals were taken every 4 h, beginning at ZT0. After RNA extraction, DNA was removed by RNase-free DNase, and 1 μg total RNA for each sample was used for making cDNA by reverse transcription (iScript cDNA kit, Bio-Rad Laboratories). cDNA was diluted 1:20 in nuclease-free water for qRT-PCR.
Based on A. burtoni sequences, qRT-PCR primers for Opn4, VA opsin, PACAP, GCO, and 2 housekeeping genes, β-tubulin and 18SrRNA, were designed (Vector NT, Invitrogen) (A. burtoni GCO: AY660539, A. burtoni rhodopsin: AF315354). Primers were designed to avoid dimers or hairpin structures, to have similar melting temperatures (ca. 60 °C), and to generate amplicons with similar length (ca. 150-200 bps).
qRT-PCR was performed in 30-μL duplicate reactions with 1 χ IQ SYBR Green Supermix (Bio-Rad Laboratories), 0.25 μM of each primer, and 2.5 ng/μL cDNA (RNA equivalent). PCR parameters were as follows: 5 min at 95 °C followed by 40 cycles of 30 sec at 95 °C, 30 sec at 60 °C, and 30 sec at 72 °C, followed by melt curve analysis. Each amplicon was specifically amplified, as demonstrated by single peaks in melt curves for each reaction product.
qRT-PCR data from MyIQ were processed using specialized software (Miner; http://www.miner.ewindup.info/) (Zhao and Fernald, 2005) to calculate cDNA concentration. To normalize, target gene concentration for each sample was divided by the geometric mean of β-tubulin and 18SrRNA concentration values. Normalized opsin mRNA levels were compared using 1-way analysis of variance (ANOVA; SPSS 11.0, SPSS Inc., Chicago, IL).
REFERENCES
- Bellingham J, Whitmore D, Philp AR, Wells DJ, Foster RG. Zebrafish melanopsin: Isolation, tissue localisation and phylogenetic position. Brain Res Mol Brain Res. 2002;107:128–136. doi: 10.1016/s0169-328x(02)00454-0. [DOI] [PubMed] [Google Scholar]
- Drivenes O, Soviknes AM, Ebbesson LO, Fjose A, Seo HC, Helvik JV. Isolation and characterization of two teleost melanopsin genes and their differential expression within the inner retina and brain. J Comp Neurol. 2003;456:84–93. doi: 10.1002/cne.10523. [DOI] [PubMed] [Google Scholar]
- Fernald RD. Retinal projections in the African cichlid fish, Haplochromis burtoni. J Comp Neurol. 1982;206:379–389. doi: 10.1002/cne.902060406. [DOI] [PubMed] [Google Scholar]
- Hannibal J. Roles of PACAP-containing retinal ganglion cells in circadian timing. Int Rev Cytol. 2006;251:1–39. doi: 10.1016/S0074-7696(06)51001-0. [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: Architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins A, Munoz M, Tarttelin EE, Bellingham J, Foster RG, Hankins MW. VA opsin, melanopsin, and an inherent light response within retinal interneurons. Curr Biol. 2003;13:1269–1278. doi: 10.1016/s0960-9822(03)00509-8. [DOI] [PubMed] [Google Scholar]
- Kojima D, Mano H, Fukada Y. Vertebrate ancient-long opsin: A green-sensitive photoreceptive molecule present in zebrafish deep brain and retinal horizontal cells. J Neurosci. 2000;20:2845–2851. doi: 10.1523/JNEUROSCI.20-08-02845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenbrot JI, Fernald RD. Circadian rhythm and light regulate opsin mRNA in rod photoreceptors. Nature. 1989;337:454–457. doi: 10.1038/337454a0. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Minamoto T, Shimizu I. A novel isoform of vertebrate ancient opsin in a smelt fish, Plecoglossus altivelis. Biochem Biophys Res Commun. 2002;290:280–286. doi: 10.1006/bbrc.2001.6186. [DOI] [PubMed] [Google Scholar]
- Moutsaki P, Bellingham J, Soni BG, David-Gray ZK, Foster RG. Sequence, genomic structure and tissue expression of carp (Cyprinus carpio L.) vertebrate ancient (VA) opsin. FEBS Lett. 2000;473:316–322. doi: 10.1016/s0014-5793(00)01550-7. [DOI] [PubMed] [Google Scholar]
- Park K, Kang HM. Circadian expression of clock genes in the rat eye and brain. Mol Cells. 2006;22:2:85–290. [PubMed] [Google Scholar]
- Philp AR, Garcia-Fernandez JM, Soni BG, Lucas RJ, Bellingham J, Foster RG. Vertebrate ancient (VA) opsin and extraretinal photoreception in the Atlantic salmon (Salmo salar) J Exp Biol. 2000;203:1925–1936. doi: 10.1242/jeb.203.12.1925. [DOI] [PubMed] [Google Scholar]
- Soni BG, Foster RG. A novel and ancient vertebrate opsin. FEBS Lett. 1997;406:279–283. doi: 10.1016/s0014-5793(97)00287-1. [DOI] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]