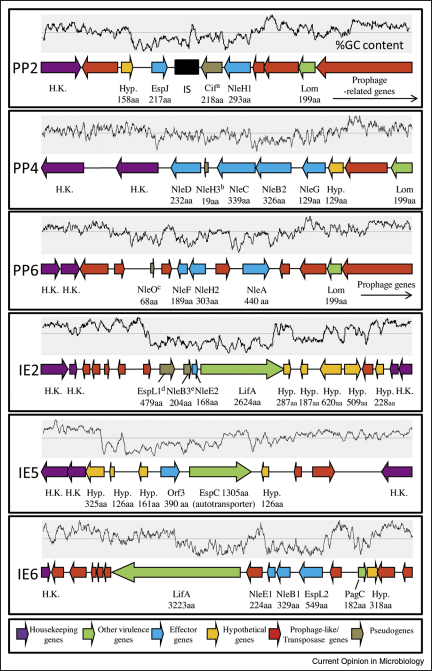
Figure 3.
The six identified non-LEE effector encoding pathogenicity islands of EPEC E2348/69. Predicted effector genes were identified by mining the EPEC genome using over 400 known/predicted effector sequences. The identified effectors and genomic islands support the genome sequence published data (see text), from which the genomic island names were obtained. Only those genomic regions encoding the effectors and with low %GC content (graph above each island) are shown with most prophage-related genes surrounding these regions omitted. Genes and strand direction are shown by individual arrows which are drawn to scale within each island and colour coded (see inset). Multiple copies of genes are numbered according to close homologues in EHEC as explained in the legend to Table 1. Pseudogene key: (a) Cif; C-terminally truncated protein not produced or secreted in this EPEC strain [48]; (b) NleH3; C-terminal truncated; (c) NleO; no start codon; (d) EspL1; stop codon in middle of gene; (e) NleB3; N-terminal truncated.