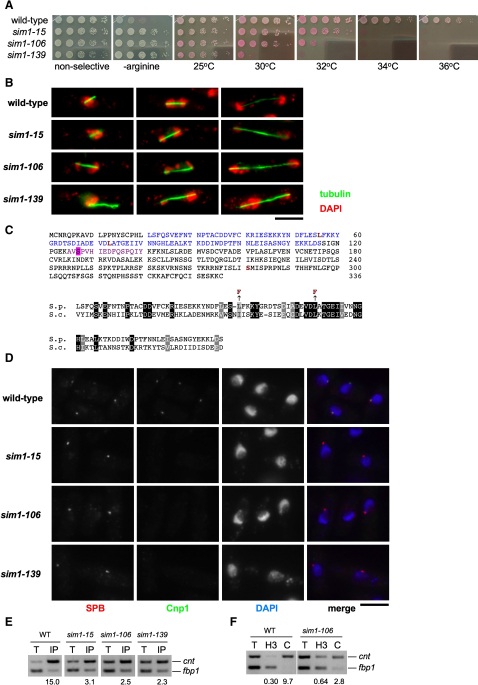
Figure 1.
sim1 Mutants Are Defective in Centromeric Silencing, Chromosome Segregation, and CENP-ACnp1 Association with the Centromere
(A) Five-fold dilutions of the indicated strains were spotted on nonselective or medium lacking arginine to assay for alleviation of silencing at the central core (cnt1:arg3+). To assess temperature sensitivity, strains were spotted onto YES + phloxine B at the indicated temperatures; dead cells stain dark pink.
(B) Chromosome segregation defects in sim1 mutants. Cells were shifted to the restrictive temperature of 36°C for 6 hr, before fixation and processing for immunofluorescence with anti-tubulin (TAT1) antibodies (green) and DAPI staining (red). Spindle length indicates that cells on left in wild-type and sim1-15 are in metaphase, and all other cells are in anaphase. Scale bar, 5 μm.
(C) S. pombe Sim1 is similar to S. cerevisiae Scm3. S. pombe Sim1/Scm3 (SPAPB1A10.02) is predicted to be 37.6 kDa. Blue: equivalent to region of Scm3Sc that binds CENP-ACse4 (Mizuguchi et al., 2007). Red: amino acid changes in sim1-139 (L56F), sim1-106 (L73F), and sim1-15 (S281L). Violet: phosphorylated tryptic peptide detected by LC MS/MS at S127 (Figure S12). Bottom: Alignment of conserved region within the CENP-ACse4 binding domain of S. cerevisiae Scm3Sc (aa 74–169) with residues 21–116 of S. pombe Scm3Sp using Clustal W/BoxShade. Amino acid changes in sim1-139 (L56F) and sim1-106 (L73F) are indicated.
(D) CENP-ACnp1 localization in scm3 mutants. Cells were shifted to 36°C for 6 hr, before fixation and processing for immunofluorescence with Sad1 antibodies to decorate spindle pole bodies (SPB; red) and provide a marker for approximate position of centromeres, labeled with anti-Cnp1/CENP-ACnp1 antisera (Cnp1; green). DAPI (blue). Scale bar, 5 μm.
(E) ChIP for CENP-ACnp1 in the indicated strains, incubated at 36°C for 6 hr prior to fixation. Primer pairs specific for central core (cnt) and a euchromatic control locus (fbp1) were used in multiplex PCR and used to calculate enrichment in IP relative to total input chromatin (T).
(F) ChIP for histone H3 (H3) and CENP-ACnp1 (C) and in wild-type and sim1-106. Primers and quantification as in (E).