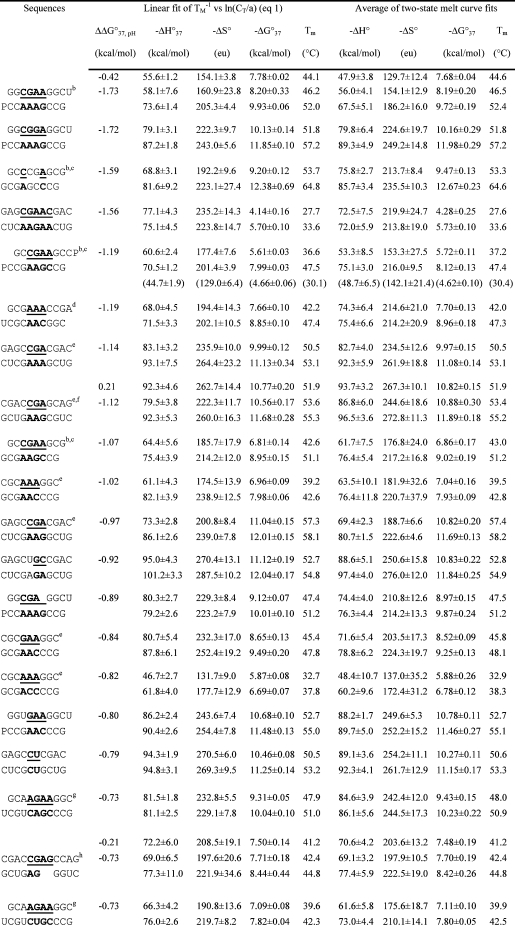
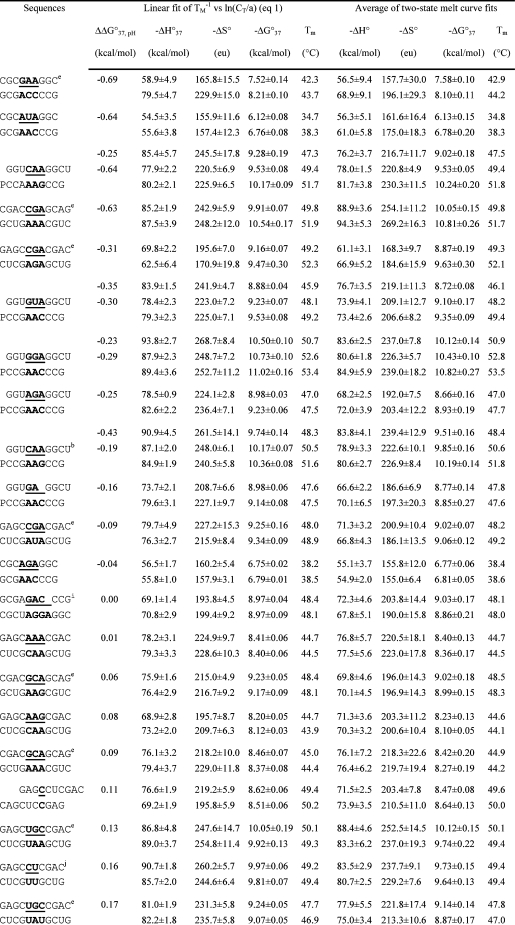
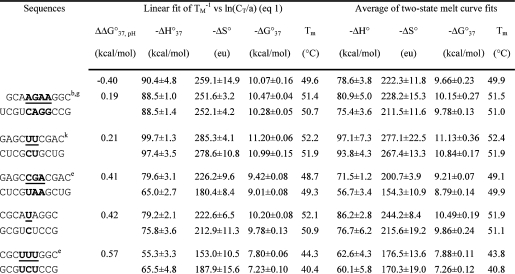
Table 1. Measured Thermodynamic Parameters for RNA Duplex Formation in 1 M NaCl.
|
For each duplex, the values from bottom to top are measured at pH 5.5, 7, and 8, respectively. Sequences are ordered from the most negative to the most positive values of ΔΔG°37,pH = ΔG°37,pH5.5 − ΔG°37,pH7, unless noted in footnote . Tm values were calculated from eq 1 at CT = 0.1 mM. Data in parentheses were measured in NMR buffer with 80 mM NaCl at pH 7.
Imino proton NMR spectra were measured (Figure 2).
ΔΔG°37,pH is per CA pair.
Loop sequence from a J4/5 loop of a group I intron (36).
Data at pH 7 are from ref (19).
Loop sequence derived from the loop A of hairpin ribozyme (3).
Loop sequence from the Alu domain of human SRP RNA (71).
The pH-independent thermodynamics is consistent with the NMR structure without the formation of the C+U pair (61).
The pH-independent thermodynamics is consistent with the NMR structure without the formation of the UC+ pair (62).
