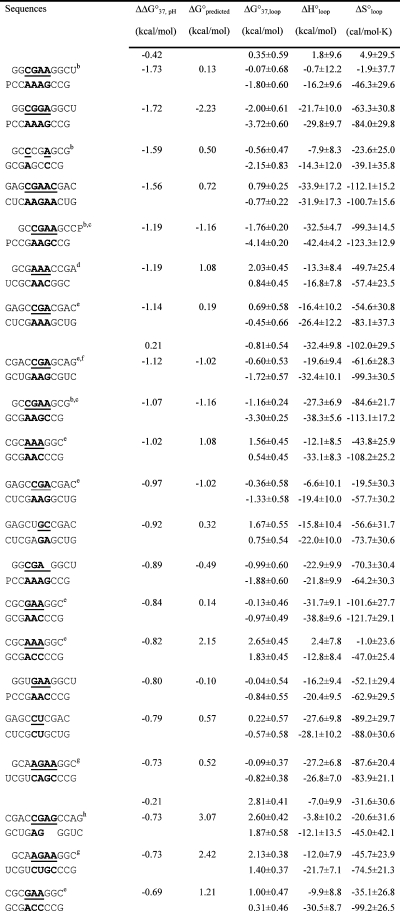
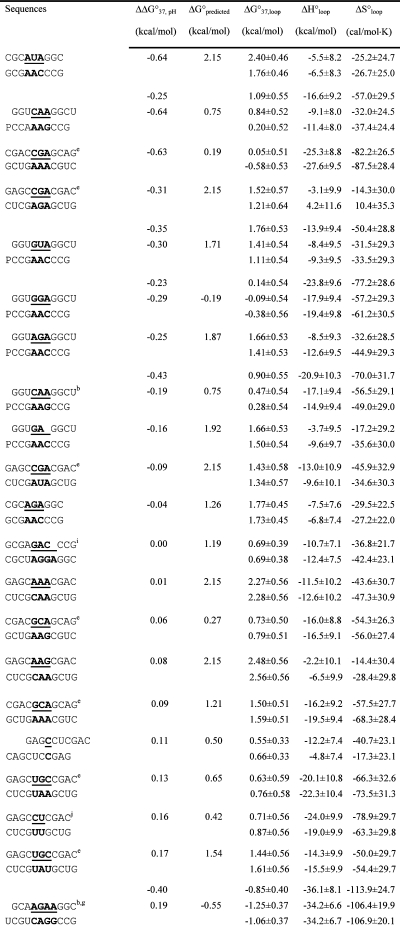
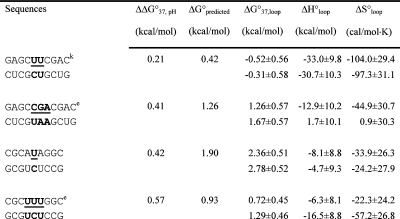
Table 2. Measured and Predicted Thermodynamic Parameters for RNA Internal Loop Formation in 1 M NaCl.
|
Calculated from eq 3a and data in Table 1 unless noted otherwise. Experimental errors for ΔG°37, ΔH°, and ΔS° for the canonical stems are estimated as 4, 12, and 13.5%, respectively, according to ref (25). These errors were propagated to estimate errors in loop thermodynamics. For each duplex, the values from the bottom to the top are measured at pH 5.5, 7, and 8, respectively. Sequences are ordered from the most negative to the most positive values of ΔΔG°37,pH = ΔG°37,pH5.5 − ΔG°37,pH7, except for (GCCCGAGCG)2 and those noted in footnote , where ΔΔG°37,pH is divided by 2. ΔG°predicted values are calculated according to eq 4. Loops smaller than 3 × 3 nucleotides are predicted according to refs (16, 29, and 31).
Imino proton NMR spectra were measured (Figure 2).
ΔG°5′CR/3′AA bonus is applied twice to predict the free energy for loop formation.
Loop sequence from a J4/5 loop of a group I intron (36).
Data at pH 7 are from ref (19).
Loop sequence derived from the loop A of hairpin ribozyme (8).
Loop sequence from the Alu domain of human SRP RNA (71).
The pH-independent thermodynamics is consistent with the NMR structure without the formation of the C+U pair (61).
The pH-independent thermodynamics is consistent with the NMR structure without the formation of the UC+ pair (62).
