Abstract
Members of the CDM (CED-5, Dock180, Myoblast city) superfamily of guanine nucleotide exchange factors function in diverse processes that include cell migration and myoblast fusion. Previous studies have shown that the SH3, DHR1 and DHR2 domains of Myoblast city (MBC) are essential for it to direct myoblast fusion in the Drosophila embryo, while the conserved DCrk-binding proline rich region is expendable. Herein, we describe the isolation of Drosophila ELMO/CED-12, an ~82 kD protein with a pleckstrin homology (PH) and proline-rich domain, by interaction with the MBC SH3 domain. Mass spectrometry confirms the presence of an MBC/ELMO complex within the embryonic musculature at the time of myoblast fusion and embryos maternally and/or zygotically mutant for elmo exhibit defects in myoblast fusion. Overexpression of MBC and ELMO in the embryonic mesoderm causes defects in myoblast fusion reminiscent of those seen with constitutively-activated Rac1, supporting the previous finding that both the absence of and an excess of Rac activity are deleterious to myoblast fusion. Overexpression of MBC and ELMO/CED-12 in the eye causes perturbations in ommatidial organization that are suppressed by mutations in Rac1 and Rac2, demonstrating genetically that MBC and ELMO/CED-12 cooperate to activate these small GTPases in Drosophila.
Keywords: Drosophila, ELMO, MBC, Rac, myoblast fusion, ommatidial organization
INTRODUCTION
Drosophila myoblast city (MBC), C. elegans CED-5, and vertebrate DOCK180, are closely related members of the evolutionarily conserved CDM family of proteins (Erickson et al., 1997; Hasegawa et al., 1996; Wu and Horvitz, 1998b). They serve as key players in a signaling complex that includes the SH2-SH3 domain-containing adaptor protein CrkII/CED-2 and the PH domain containing protein ELMO/CED-12 (reviewed in (Meller et al., 2005)). This complex then acts at the membrane to relay signals to the small GTPase Rac1/CED-10. MBC/DOCK180/CED-5 function as non-conventional Guanine nucleotide Exchange Factors (GEFs) for Rac. Conventional GEFs bind to nucleotide-free Rac via a Dbl-homology (DH) domain, thereby facilitating exchange of GDP for GTP. DOCK180/CED-5, which lack DH domains, associate with nucleotide-free Rac through a conserved Dock-Homology Region (DHR2) (Brugnera et al., 2002; Cote and Vuori, 2002). Deletion of this domain results in loss of Rac binding and the inability to direct formation of GTP-bound Rac (Brugnera et al., 2002).
In addition to DHR2, CDM proteins have in common an N-terminal SH3 domain, a second Dock Homology Region (DHR1), and a C-terminal proline rich region. The C-terminal region directs interaction with the SH3 domain of CrkII/CED-2. The CrkII SH2 domain can then direct interaction with upstream proteins that are phosphorylated on tyrosine, such as transmembrane receptors and components of focal adhesions (Cheresh et al., 1999; Kiyokawa et al., 1998). In addition to membrane recruitment through Crk-related interactions, the C-terminal PH domain of ELMO/CED-12 can also mediate its membrane localization (deBakker et al., 2004; Grimsley et al., 2004). The DHR1 region of DOCK180, which binds to phosphatidylinositol 3,4,5-triphosphate [PtdIns(3,4,5)P3], is also required for its membrane localization (Cote et al., 2005; Kobayashi et al., 2001). The N-terminal SH3 domains of DOCK180 and CED-5 mediate interaction with the C-terminal proline-rich region of ELMO and CED-12, respectively (Lu et al., 2004; Lu et al., 2005; Wu et al., 2001). In vitro studies demonstrate that DOCK180 binding to Rac can be sufficient for its activation (Cote et al., 2005), but that this activation can be significantly enhanced by DOCK180 bound to ELMO (Brugnera et al., 2002; Katoh and Negishi, 2003; Lu et al., 2004). Thus, the DOCK180/ELMO complex is a key component in CDM signaling to Rac.
In addition to extensive homology and conserved biochemical interactions between the DOCK180/CED-5 and ELMO/CED-12 protein families, complexes of these proteins perform similar biological functions. For example, genetic studies have shown that C. elegans CED-10 acts with CED-2/Crk, CED-5/DOCK180 and CED-12/ELMO to promote cell migration of the distal tip cells during development of the somatic gonad and engulfment of cell corpses following apoptosis (Gumienny et al., 2001; Kinchen et al., 2005; Wu and Horvitz, 1998a; Zhou et al., 2001). Membrane targeted Dock180 increases cell spreading, and overexpression of wild-type Dock180 in mammalian cells enhances cell migration and phagocytosis of apoptotic cells (Cheresh et al., 1999; Kiyokawa et al., 1998). Lastly, a reduction in wild-type Dock180 or overexpression of mutant forms of DOCK180 decrease activated Rac and cause defects in cell spreading and cell migration (Cote and Vuori, 2002; Katoh and Negishi, 2003; Kiyokawa et al., 1998).
As in C. elegans and vertebrates, Drosophila MBC interacts genetically with other molecules required for CDM pathway function. For example, mutations in mbc delay border cell migration and influence PVR mediated F-actin accumulation in the follicle cells during ovary development, reflecting its proposed role in the PVR-Rac pathway (Duchek et al., 2001). Studies utilizing RNAi constructs have demonstrated a genetic interaction between MBC, Drosophila Crk (DCrk) and ELMO in adult thorax closure (Ishimaru et al., 2004). In the Drosophila eye, in which Rac is required for proper actin organization (Chang and Ready, 2000), the effects of Rac1 overexpression are suppressed by loss of one copy of mbc (Nolan et al., 1998). Rac1 was initially implicated in myoblast fusion in the embryo by the demonstration that overexpression of either dominant-negative or constitutively-active Rac1 interferes with myoblast fusion (Luo et al., 1994). More recently, loss-of-function studies have established that Rac1 and Rac2 play a redundant but essential role in this process. MBC is absolutely essential for myoblast fusion (Hakeda-Suzuki et al., 2002), where it colocalizes in founder myoblasts with the immunoglobulin superfamily member Duf/Kirre (Chen and Olson, 2001). MBC is also expressed in, and apparently required in, the fusion competent myoblasts (Balagopalan et al., 2006). Structure/function analysis of MBC identified the protein domains that are essential for myoblast fusion. This analysis revealed that, like Dock180, the DHR1 domain binds to PtdIns(3,4,5)P3 and is essential for myoblast fusion. The N-terminal SH3 domain and the putative Rac binding DHR2 domain are essential for MBC transgenes to rescue the myoblast fusion defects in mbc mutant embryos. Surprisingly, however, while the C-terminal proline-rich region directs a strong interaction with DCrk (Balagopalan et al., 2006; Ishimaru et al., 2004), this region is not required for MBC function in the embryonic musculature(Balagopalan et al., 2006). Thus, the canonical Crk-associated CDM pathway that has been described for other organisms and used in other Drosophila tissues (Bourne, 2005; Ishimaru et al., 2004; Reif and Cyster, 2002) does not appear to be used in Drosophila myoblast fusion. This finding raised the broader question of whether other components of the canonical pathway functioned along with MBC in the embryonic muscle, or whether MBC functions in this tissue through different protein interactions.
To address the involvement of Drosophila ELMO in signaling from MBC, we have isolated and characterized its role in myoblast fusion, border cell migration and ommatidial organization. Herein we demonstrate that ELMO is expressed in the ovary, where it interacts genetically with MBC and plays a role in border cell migration. We generate and analyze multiple loss-of-function alleles to demonstrate the importance of maternal and zygotic ELMO in myoblast fusion. We confirm, through a targeted mass spectrometry approach, that these proteins form a stable complex in the embryonic musculature. We demonstrate that ELMO and MBC act cooperatively in the mesoderm, such that an excess of both causes serious defects in myoblast fusion reminiscent of those seen with excess Rac activity. Finally, we establish genetically that ELMO and MBC can cooperate to activate Rac GTPases in the adult eye.
MATERIALS AND METHODS
Drosophila stocks
Fly stocks were raised on standard cornmeal medium at 25°C unless otherwise indicated. Oregon R was used as the wild-type strain. Fly stocks that were obtained from the Bloomington Stock Center include UAS-mCD8-GFP, c306-GAL4, UAS-RacN17, PBac[c06760]/CyO, and Df(2L)Prl/CyO. The UAS-elmoIR, UAS-mbcIR and UAS-DCrkIR transgenic lines were obtained from Ryu Ueda (Ishimaru et al., 2004). UAS-elmo and UAS-elmoHA were generated by standard methods and injected by Genetic Services, Inc. UAS-mbc and UAS-mbcHA have been described (Balagopalan et al., 2006). For analysis of the border cell migration phenotype, slbo-Gal4; UAS-mCD8-GFP or c306-Gal4 flies were crossed to UAS transgenic flies. Non balancer progeny were fattened overnight at 29°C before dissection.
Molecular cloning of elmo
A full length cDNA of the elmo transcript was obtained by RT-PCR from Oregon R flies using standard methods with the forward primer GCGCCCGGGAATGATACCAAAAAAGACGACGG and the reverse primer CGCGCTCGAGTTAGCTCTCAAAGCAAAAATCATAG. The resulting PCR product was digested and cloned into the yeast 2-hybrid prey vector pGADT7 (Clontech). Full length elmo was amplified by PCR and cloned into pUAST using the forward primer ATAAGAATGCGGCCGCAATGATACCAAAAAAGACGACGG and the reverse primer GCTCTAGAGTTAGCTCTCAAAGCAAAAATC. For the HA-tagged UAS-elmo construct, sequences encoding the HA epitope were added to the 3′ end. All clones were verified by sequencing.
In situ hybridization and immunohistochemistry
Embryos were collected on agar-apple juice plates and aged at 25°C. For in situ analysis, full-length elmo was transcribed with Sp6 using the DIG mRNA labeling kit (Roche) and hybridized as described (Tautz and Pfeifle, 1989). Myosin heavy chain was detected colorimetrically as described (Erickson et al., 1997)
Ovary dissections and immunofluorescence staining were performed as in Geisbrecht et al. (Geisbrecht and Montell, 2004). Antisera to ELMO was generated in rabbit using full length protein overexpressed in bacteria and used at 1:10. It was detected fluorescently using Alexa Fluor® 488 goat anti-rabbit IgG at 1:400 (Molecular Probes, Carlsbad, CA). 0.5 mg/ml DAPI (Sigma, Dallas, TX) and Alexa Fluor® 546-phalloidin at 1:400 (Molecular Probes, Carlsbad, CA) were added with the secondary antibodies.
Yeast 2-hybrid assays
The yeast 2-hybrid screen was performed by PanBionet Inc. Full length elmo cDNA was cloned as above and yeast 2-hybrid experiments to confirm this interaction were performed as in Balagopalan, et al. (Balagopalan et al., 2006).
Mass spectrometry
For both MBC and ELMO, HA-tagged and untagged transgenic flies were crossed to mef2-GAL4 females and 6–18h embryos were collected on agar-apple juice plates at 25°C. For each immunopreicitation experiment, ~300 mg embryos were dechorionated and homogenized in lysis buffer [60mM Tris (pH 7.5), 80mM NaCl, 6mM EDTA (pH 8.0), 2% Triton X-100, 1mM Na3VO4, 5mM 1-Naphthyl phosphate potassium salt, 2mM PMSF, 2 ug/ml Leupeptin, 2 ug/ml Pepstatin]. The NaCl concentration was increased to 300mM and resulting lysate mixed with anti-HA resin overnight at 4°C. The resin was washed 3 times with wash buffer [60mM Tris (pH 7.5), 300mM NaCl, 6mM EDTA (pH 8.0), 1.0% Triton X-100] with protease inhibitors. Protein was eluted from the resin using 0.2 mg/ml 2x HA peptide. Samples were TCA-precipitated and digested as described (Florens and Washburn, 2006). Peptide digests were separated on 3-phase (reverse-phase/strong cation exchange/reverse phase) chromatography microcapillary columns(McDonald et al., 2002), and analyzed by Multidimensional Protein Identification Technology (MudPIT) (Washburn et al., 2001). Tandem mass (MS/MS) spectra were matched using SEQUEST (Eng et al., 1994) to peptides from a database of 35050 amino acid sequences, consisting of 17348 D. melanogaster proteins (non-redundant entries from NCBI 2006-11-28 release) and, to estimate false discovery rates, 17525 randomized sequences for each non-redundant protein entry. Peptide/spectrum matches were selected and compared using DTASelect/CONTRAST (Tabb et al., 2002) and were retained if they had a DeltCn of at least 0.08. Peptides had to be at least 7 amino acids long and their termini had to comply with the proteolytic enzyme selectivities. For trypsin digests, minimum cross-correlation scores (Xcorr) were set at 1.8 for singly-, 2.0 for doubly-, and 3.0 for triply-charged spectra, while for elastase digests, minimum XCorr were 2.5 for doubly-, and 3.5 for triply-charged spectra. Combining all runs, proteins had to be detected by at least 2 such peptides, or 1 peptide with 2 independent spectra. To estimate relative protein levels, Normalized Spectral Abundance Factors (NSAFs) were calculated for each non-redundant protein, as described in (Paoletti et al., 2006; Zybailov et al., 2006)
Generation and molecular characterization of elmo alleles
Isogenized b1elA1rdspr1 cn1or49h (Bloomington Stock Center) males were treated with 25mM EMS in 5% sucrose for 18–24h using standard methods, and the mutagenized males mated with Gla/CyO, GAL4-twi, P{UAS-2xEGFP] virgin females. Approximately 5000 CyO-balanced male progeny were mated individually to PBac[c06760]/CyO females. Stable stocks were generated from crosses that failed to complement, and balanced with Gla/CyO, GAL4-twi, P{UAS-2xEGFP]. For each EMS allele, the elmo genomic region was amplified by PCR from heterozygous flies using elmo-specific primers and the PCR products sequenced to identify the molecular lesion. All mutations were confirmed in multiple sequencing reactions from independently amplified DNA samples. For RT-PCR, RNA from appropriately staged material was made using Trizol and subsequent RT-PCR was performed using elmo-specific primers by standard methods. Df(2L)HO55 was made by P-element excision of the Bloomington stock 10539 in the downstream gene aats-thr. Removal of elmo was confirmed by PCR.
Generation of germ line and mosaic clones and RNAi analysis
The elmoPB, elmo8C6, and elmo19F3 mutations were recombined onto the FLP recombination target (FRT)-containing second chromosome of w; al1 dp ov1 b1 pr1 P{ry[+t7.2]=neoFRT}40A (Bloomington Stock Center) and mated to males of the genotype hs-FLP22; P{ry[+t7.2]=neoFRT}40A, P{ovoD1}/CyO. Progeny from this cross were heat shocked at 37°C for 2h on days 4 and day 5 (late L2 or L3) as described (Balagopalan et al., 2006). Female progeny of the genotype hs-FLP22/+; P{ry[+t7.2]=neoFRT}40A, P{ovoD1}/P{ry[+t7.2]=neoFRT}40A, elmo* were recovered and crossed to Df(2L)Prl/CyO, GAL4-twi, P{UAS-2xEGFP] heterozygous males. The muscle pattern of embryos laid by these females was then examined as described above. For mosaic clonal analysis, larvae of the genotype hs-FLP22/+; P{ry[+t7.2]=neoFRT}40A, P{ovoD1}/P{ry[+t7.2]=neoFRT}40A, elmo19F3, ubiGFP were heat shocked as described (Niewiadomska et al., 1999). The ovaries of female progeny of the appropriate genotype were then dissected and examined as described above. For RNAi analysis in embryos, the progeny of mef2-GAL4 or mef2-GAL4/2x UAS-elmoIR were grown at, 29°C until stage 16, immunostained with MHC as above, and unfused myoblasts were counted in hemisegments A2-A5.
Scanning Electron Microscope (SEM) analysis
Scanning electron micrographs of the eyes of adult flies of the correct genotype were taken using the Hitachi Tabletop Microscope TM-1000 according to manufacturer’s suggestions.
RESULTS
Identification of ELMO as a binding partner for MBC in the Drosophila musculature
The SH3 domain of MBC is required during Drosophila muscle development (Balagopalan et al., 2006). We therefore used this domain as bait in a yeast two-hybrid screen of a Drosophila embryonic cDNA library to identify binding partners that may function with MBC in myoblast fusion. This screen resulted in the identification of CG5336, the Drosophila ortholog of ELMO/CED-12. The interaction between full length MBC, or MBC that lacks all proline rich regions, and full length ELMO was confirmed in a yeast two-hybrid assay (Fig. 1A). It was also confirmed by immunoprecipitation of lysate from S2 cells co-transfected with full length HA-tagged MBC and Flag-tagged ELMO in an analysis similar to that of Ishimaru et al. (Ishimaru et al., 2004)(data not shown).
Figure 1.
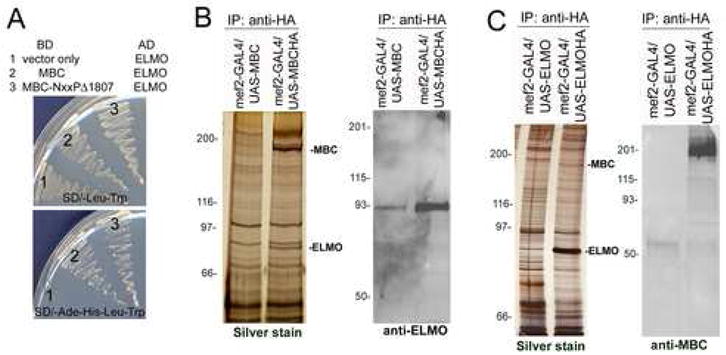
Identification of ELMO as a binding partner for MBC. (A) Yeast two-hybrid assay. Growth on selective media is shown on the lower panel where both full-length and truncated MBC exhibit interaction with ELMO. (B, C) Anti-HA immunoprecipitates from lysates of untagged and HA-tagged MBC (B) and ELMO (C) expressed in the Drosophila mesoderm under control of mef2-GAL4. (B) The silver stained gel (left panel) shows a band of ~83kD that co-precipitates with MBC-HA. This band is revealed as ELMO by mass spectrometry and Western blotting with an antisera against ELMO (right panel). (C) The silver stained gel (left panel) shows a band of ~200kD that co-precipitates with ELMO-HA. This band is identified as MBC by mass spectrometry and Western blotting with an antisera against MBC (right panel).
In previous studies, similar assays had revealed a robust interaction between MBC and DCrk that is not required for MBC-mediated signaling in myoblast fusion (Balagopalan et al., 2006). Thus, as a first step in determining the relevance of the above MBC/ELMO complex to the embryonic musculature, we chose to use a proteomics analysis of MBC interactions in this tissue. Flies transgenic for either HA-tagged or untagged MBC under UAS control were crossed to flies bearing the muscle-specific mef2-GAL4 driver, and lysates prepared from the resulting embryos at approximately 6–18 h AEL. Both transgenes are functional and rescue the myoblast fusion phenotype when expressed in the mesoderm of mbc mutant embryos (Balagopalan et al., 2006). HA-tagged MBC and associated proteins were then immunoprecipitated with anti-HA resin, and proteins in the immunoprecipitate analyzed in a silver stained gel, by western blot and by Multidimensional Protein Identification Technology (MudPIT) mass spectrometry (Washburn et al., 2001). In control samples, lysate from untagged MBC was subjected to identical immunoprecipitation and analysis.
As shown in Figure 1B, a band corresponding to the expected molecular weight of ELMO was detected in a silver stained gel of total protein immunoprecipitated from lysates containing HA-tagged MBC but not untagged MBC. Immunobloting confirmed this result, detecting a strong band in the immunoprecipitate from HA-tagged MBC. A small amount of ELMO was also present in the control immunoprecipitate, and appeared to be due to nonspecific sticking of MBC to the HA-resin. In the proteomic analysis of the immunoprecipitates, shown in Table 1, ELMO was the most abundant protein detected in all MBC-HA runs aside from MBC itself. To further substantiate this interaction, reciprocal immunoprecipitations utilized embryos in which HA-tagged or untagged ELMO transgenes were expressed in the mesoderm under mef2-GAL4 control. While the presence of MBC was not as apparent in total immunoprecipitated protein analyzed by silver staining, immunoblotting revealed a strong band that cross reacted with antisera against MBC. In samples analyzed by mass spectrometry, MBC was detected in all HA-ELMO immunoprecipitates, with spectral counts corresponding to the most abundant peptides detected after ELMO (Table 1). As anticipated from our previous studies, DCrk was not detected in samples of immunoprecipitated MBC or ELMO analyzed by MudPIT, nor were these proteins detected in samples of HA-tagged DCrk expressed in the embryonic musculature.
Table 1.
Identification of Interacting proteins by MudPIT Analysis
| MBC-interacting Proteins | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein | Ti_1 | Ti_2 | Ti_3 | Ti_4 | Ti_5 | Ti_6 | Es_1 | EC_1 | Merged controls |
| MBC | 27.7%a(164)b | 4.4% (6) | 28.8% (202) | 49.2% (448) | 5.3% (10) | 11.6% (30) | 11.2% (18) | 6.2% (16) | 1.7% (2) |
| CED-12 | 26.9% (61) | 5.4% (4) | 33.0% (84) | 45.3% (131) | 1.8% (1) | 11.3% (9) | 2.1% (1) | 0.0% (1) | 0.0% (0) |
| CRK | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) |
|
| |||||||||
|
CED-12 interacting Proteins | |||||||||
| Protein | Ti_1 | Ti_2 | Ti_3 | Ti_4 | Ti_5 | Es_1 | Es_2 | Es_3 | Merged controls |
|
| |||||||||
| CED-12 | 42.5%a(2043)b | 69.7% (1151) | 74.5% (1985) | 50.1% (2923) | 70.7% (2923) | 71.1% (867) | 73.8% (790) | 65.5% (432) | 1.8% (2) |
| MBC | 7.2% (89) | 27.4% (118) | 39.3% (244) | 27.9% (398) | 31.3% (245) | 25.4% (112) | 8.4% (18) | 2.3% (5) | 0.0% (0) |
| CRK | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) | 0.0% (0) |
Sequence coverage, percentage of the protein sequence covered by detected peptides identified by tandem mass spectroscopy
Spectral counts, total spectra matching peptides detected by tandem mass spectroscopy
Ti=trypsin
Es=elastase
EC=endoproteinase Glu-C
ELMO is expressed ubiquitously during oogenesis and embryogenesis
To examine the temporal expression pattern of elmo mRNA, RT-PCR was performed at various stages throughout Drosophila development (Fig. 2A). Much like mbc, the elmo transcript is abundant during oogenesis and provided maternally to the embryo, as evidenced by the presence of RT-PCR product in unfertilized eggs and embryos at 0–4 h AEL. Expression remains relatively high throughout embryogenesis, persists during the larval and pupal stages, and is detectable in the adult. The spatial expression pattern of ELMO, as revealed by antisera raised against the full length protein, is broad and cytoplasmic in the developing Drosophila ovary (Fig. 2B–C″). It is constant in the germline and becomes evident in the somatic follicle cells after stage 9 (Fig. 2B, arrow), with higher expression in the border cells (Fig. 2C inset, C′, arrow) at stage 10. In the embryo, maternally-provided elmo transcript is high prior to stage 5, which coincides with the beginning of zygotic transcription (Fig. 2D). It is present in the developing mesoderm in stages 9–13 (Fig. 2E–G, arrows) and remains detectable in both the visceral and somatic musculature through Stage 16 (Fig. 2H, I, arrows). Staining with polyclonal rabbit antisera against ELMO reveals a broad temporal and spatial pattern of expression, consistent with the results of RT-PCR and in situ hybridization (data not shown).
Figure 2.
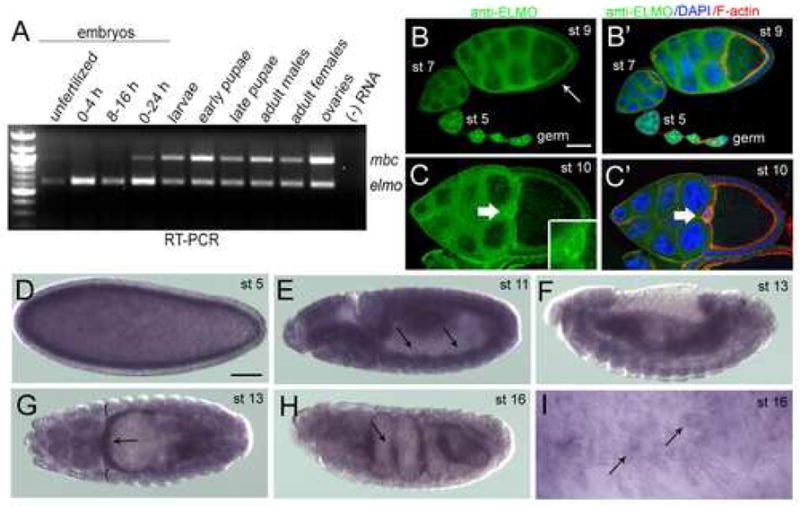
Temporal and spatial expression patterns of elmo. (A) Temporal expression of mbc and elmo transcript by RT-PCR at specific developmental stages in the Drosophila life cycle (B–C′) Immunoreactivity of ELMO antisera in wild-type ovarioles. (B) ELMO is high in the germline at all stages of ovariole development, and is detected in the somatic follicle cells around stage 9 (arrow). (B′) DAPI (blue) and F-actin (red) do not extensively overlap with ELMO (green). (C) ELMO expression is ubiquitous in both the germline and somatic follicle cells of a stage 10 egg chamber, and is especially prominent in the border cells (arrow, inset). (C′) DAPI (blue) and F-actin (red) do not overlap extensively with ELMO (green). (D–I) in situ analysis of elmo in wild-type embryos. (D) elmo is detected at stage 5, suggesting that it is provided maternally. (E) Expression is fairly ubiquitous throughout the embryo, with mesodermal expression (arrows) becoming apparent by stage 11. (F, G) Stage 13 embryo in which expression is broad, but stronger in the gut visceral mesoderm (arrows) than in the somatic mesoderm (brackets). (H) Expression is highest (arrow) in the visceral mesoderm at stage 16. (I) High magnification view of the somatic mesoderm showing elmo expression in the mature muscle. Lateral views are shown in (D–F, H–I) and a dorsal view in G. Scale bar, 50μM
Generation of EMS-induced mutations in elmo
Drosophila CG5336/ELMO has 47% overall similarity with C. elegans CED-12 and 45–54% with the three vertebrate ELMO proteins. All proteins contain the same signature domains (Fig. 3A). The primary amino acid sequence of the N terminal two thirds of ELMO shares no obvious homology with any protein domains. However, analysis of secondary structure using a homology detection/structure prediction program and psi-blast http://toolkit.tuebingen.mpg.de/hhpred) suggests that this region adopts a superhelical structure similar to that of the ARMADILLO/HEAT repeat, and includes five such repeats ((deBakker et al., 2004); Mushegian and Geisbrecht, unpublished). The PH domain is highly conserved, and all proteins contain a single proline-rich PxxP consensus sequence for binding to SH3 domains.
Figure 3.
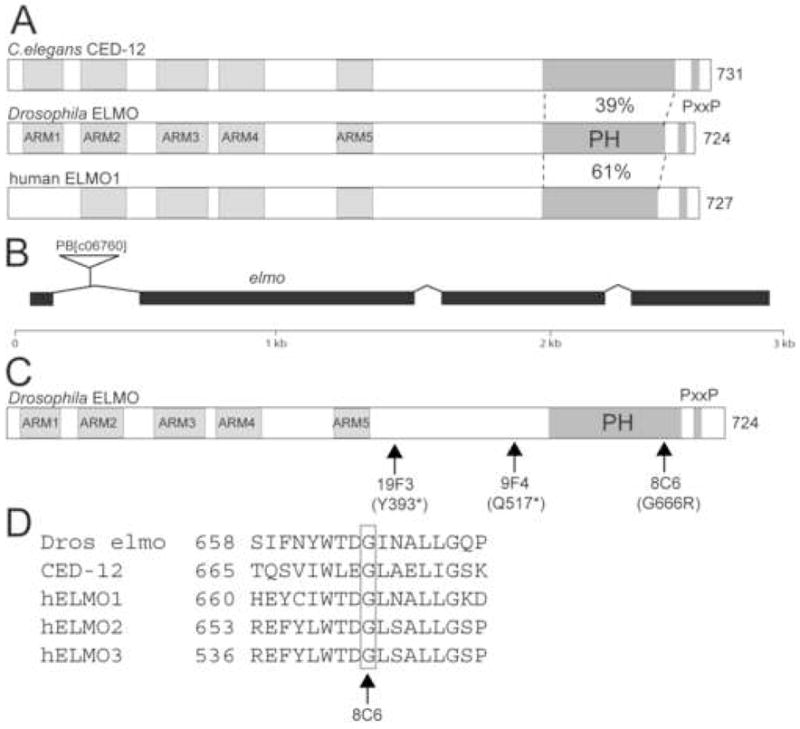
CG5336 encodes ELMO/Dced-12. (A) Drosophila ELMO is the ortholog of C. elegans CED-12 and human ELMO1. The N-termini of these proteins share predicted Armadillo repeats, while the C-terminal region contains a pleckstrin homology (PH) domain and proline-rich region. (B) The intron/exon organization of the CG5336/elmo locus is shown. The PiggyBac transposable element PB[c06760] is located in the first intron. (C) The molecular lesions obtained in an EMS screen for elmo mutants are indicated. (D) The molecular lesion in the EMS-induced allele elmo8C6 occurs in a conserved glycine at amino acid position 666 within the PH domain.
Genetic analysis of elmo was initiated using a lethal PiggyBac insertion in the first intron (PB[c06760]; Fig. 3B). The lethality of the insert is reverted by its excision or by expression of a UAS-elmo transgene under control of actin-GAL4. Molecular analysis of embryos both maternally and zygotically mutant for PB[c06760] by RT-PCR revealed the presence of wild-type transcript as well as two larger products (Supp Fig. 1). The nucleotide sequence of these larger products indicates that the PB[c06760] insertion causes some aberrant splicing of the primary transcript between exons 1 and 2, and introduces a stop codon 8 amino acids downstream from the end of exon 1. Despite this perturbation, the presence of wild-type transcript suggests that PB[c06760] is a hypomorphic allele of elmo. To obtain null alleles, we carried out an F2 lethal screen of 5000 EMS-mutagenized chromosomes for individuals that failed to complement PB[c06760]. The molecular lesions of the resulting alleles are shown in Figure 3C and D. Stop codons have been introduced before the PH domain in elmo19F3 and elmo9F4, while elmo8C6 represents a missense mutation in which a conserved glycine has been changed to arginine.
Reduced ELMO causes defects in border cell migration
Based on the observation that mbc has a role in border cell migration (Duchek et al., 2001), that ELMO is expressed in the border cells, and that ELMO interacts physically with MBC, we examined whether ELMO is also required for border cell migration. In this process, a group of 8–10 border cells are recruited from the anterior follicle cells during stage 9, and migrate between the nurse cells until they reach the nurse cell/oocyte border at stage 10 (Fig. 4A). To examine the requirement for elmo during this migration, expression of UAS-elmo RNAi was directed to the border cells using the slbo-GAL4 driver. The RNAi-mediated reduction of mbc serves as a positive control and, as expected, results in severe defects in border cell migration in which approximately 40% of the stage 10 egg chambers contain cell clusters that fail to reach the nurse cell/oocyte border (Fig. 4B). Similar, but weaker, effects are caused by an RNAi-mediated reduction in elmo. Two independent transgenic lines, each expressing one copy of UAS-elmo RNAi, cause mild defects in which 20% of the border cells fail to migrate completely (Fig. 4B). The penetrance of this defect increases to 40% in the presence of two copies of UAS-elmo RNAi. Co-expression of UAS-mbc RNAi and UAS-elmo RNAi transgenes further increases the severity and penetrance of the defects, such that 55% of the border cells fail to migrate completely. By comparison, co-expression of UAS-DCrk RNAi and UAS-mbc RNAi increases the observed defects in border cell migration only slightly over those observed for each transgene alone. Similar results were obtained when the above transgenes were driven in the border cells using c306-Gal4 driver (data not shown).
Figure 4.
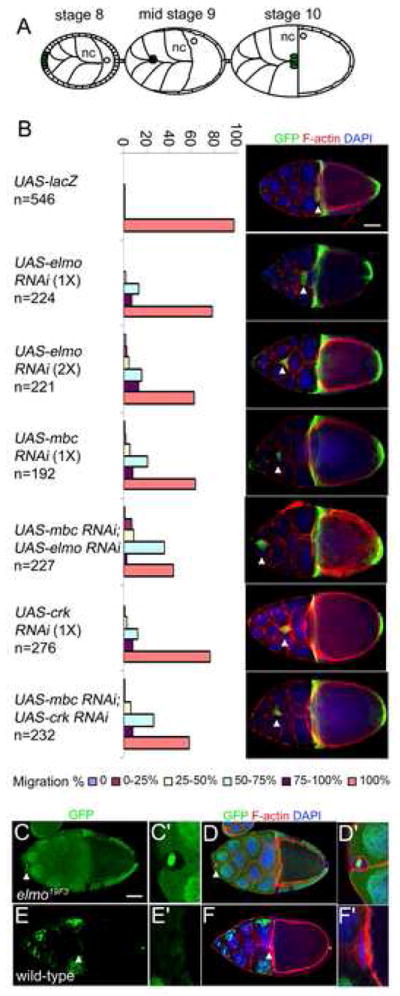
Abrogation of elmo function results in border cell migration defects.
(A) Schematic of egg chambers at stages 8–10. Nurse cells (nc), oocyte (o) and border cells (green) are indicated. (B) Quantification of border cell migration defects in stage 10 egg chambers expressing the indicated RNAi transgene. Transgene expression is driven by slbo-Gal4, in a genetic background that also includes UAS-mCD8-GFP. The number of stage 10 egg chambers examined for each genotype is provided. A confocal image of one representative stage10 egg chamber is provided for each genotype to show the migration defect. The arrowhead in each indicates the position of the border cells. Egg chambers were stained for GFP (green), F-actin (phalloidin, red) and DAPI (blue). (C–F′) Mosaic analysis of elmo19F3. Confocal images of stage 10 egg chambers of elmo19F3 (C–D) and wild-type (E–F). High magnification views of the same border cell cluster which in the elmo19F3 mutant contain a mixture of GFP (green) positive and GFP negative cells and show no migration (C′) and in wild-type are all GFP negative and show complete migration (E′). (D–D′ and F–F′) Overlap with DAPI (blue) and F-actin (phalloidin, red) do not extensively overlap with GFP (green) expression. Arrowhead indicates the position of the border cells. Scale bar, 50μM
To ensure that the RNAi effects observed in border cell migration are due to a reduction in elmo gene function, mosaic clones of border cells lacking all elmo expression were generated using the elmo19F3 stop codon allele. As described recently by others (Bianco et al., 2007), we observed severe migration defects in border cells lacking elmo with 35% showing no migration at all and the remaining 65% arresting their journey when only 25% complete (Fig. 4C–F′ and data not shown).
ELMO is required for myoblast fusion
Since the MBC SH3 domain is essential for embryonic myoblast fusion (Balagopalan et al., 2006) and this domain interacts with ELMO, we examined whether loss of elmo resulted in defects in myoblast fusion. Embryos homozygous for elmoPB[c06760] or elmo8C6, both of which are hypomorphic alleles, have minor defects in which a small number of myoblasts remain unfused and muscles are occassionally missing (data not shown). Embryos trans-heterozygous for elmoPB[c06760] or elmo8C6 and Df(2L)HO55, a deficiency that removes elmo, also exhibit very modest perturbations. The overall muscle pattern is unperturbed in these embryos, but some unfused myoblasts are present just under the muscle layer when compared to wild-type embryos in which these unfused myoblasts are not present (Fig. 5A, A′, C–D′). A larger number of unfused myoblasts as well as missing muscles are apparent in Df(2L)HO55/Df(2L)HO55 embryos (Fig. 5B, B′) and embryos trans-heterozygous for Df(2L)HO55 and either elmo9F4 or elmo19F3, the latter of which contain stop codons and are therefore likely to be protein null (Fig. 5E, F). Of note, the unfused myoblasts are most abundant and more easily visualized in the focal plane just below the muscle layer (Fig. 5E′, F′).
Figure 5.
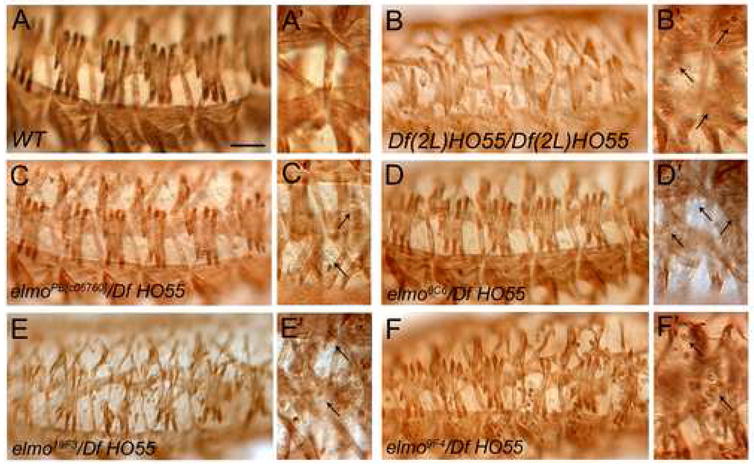
Somatic muscle phenotype of elmo zygotic mutant embryos. (A–F′) Low and high magnification views of stage 16 embryos stained immunohistochemically with anti-MHC to reveal the muscle pattern. (A) Wild-type embryos showing the mature muscle pattern and lack of unfused myoblasts (A′). (B) Embryos homozygous mutant for Df(2L)HO55, a deficiency that removes elmo, exhibit missing muscles and unfused myoblasts (B′). (C, D) The muscle pattern appears to be unperturbed in transheterozygous embryos zygotically mutant for elmoPB[c06760]/Df HO55 (C) and elmo8C6/Df HO55 (D), with a few unfused myoblasts visible under the muscle layer (C′, D′, arrows). (E, F) Muscle fibers are missing in transheterozygous embryos zygotically mutant for elmo19F3/Df HO55 (E) and elmo9F4/Df HO55 (F), and many unfused myoblasts are apparent (E′, F′, arrows). Scale bar, 50μM
The presence of maternally provided transcript and the variable penetrance of the embryonic muscle defects suggested that the perdurance of maternal elmo mRNA and/or its protein product may be obscuring the loss of function muscle phenotype. To address this hypothesis, germline clones (GLCs) were generated to eliminate both maternal and zygotic elmo transcripts. GLCs of elmo9F4 do not survive oogenesis, likely due to the presence of a lethal background mutation. GLC embryos mutant for elmo19F3 die prior to myogenesis, possibly reflecting an earlier embryonic requirement for elmo. We therefore examined GLC embryos maternally mutant for elmoPB[c06760] or elmo8C6 and zygotically heterozygous for Df(2L)Prl, which removes elmo, with the hope that these hypomorphic alleles would provide sufficient transcript for early embryogenesis, and allow later visualization of the musculature under conditions of reduced elmo. As shown in Figure 6A and B, these embryos exhibit a greater number of missing muscles and unfused myoblasts (Fig. 6A′, B′) compared to zygotic trans-heterozygotes of the hypomporphic alleles elmoPB[c06760] or elmo8C6. Thus, both the maternal and zygotic transcripts contribute to ELMO’s role in the embryonic musculature
Figure 6.
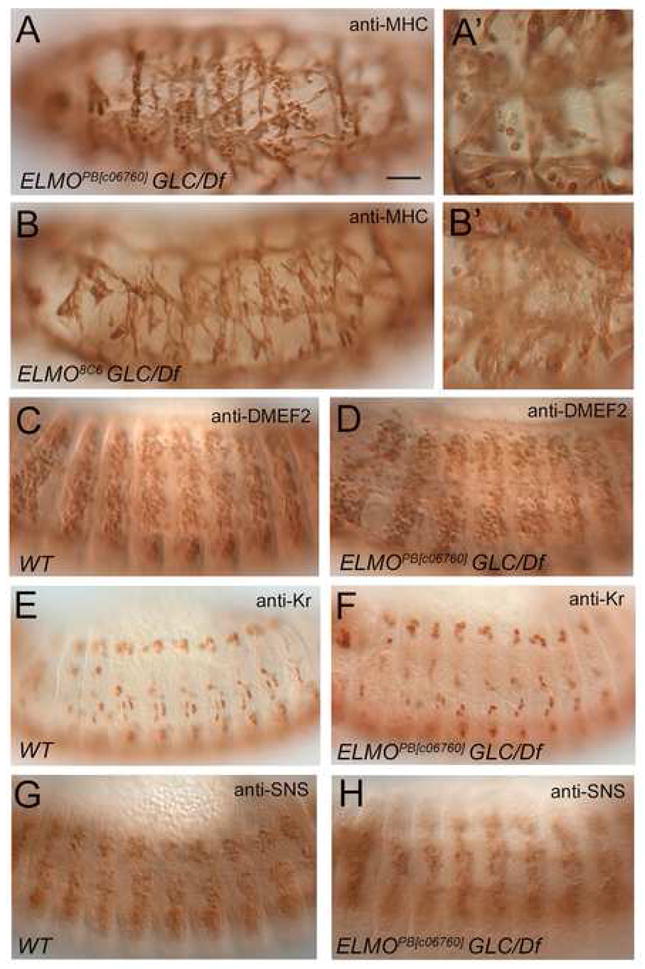
Somatic muscle phenotype and muscle cell specification of maternal/zygotic elmo mutant embryos. (A-B′) Embryos from GLCs of elmoPB[c06760] or elmo8C6, that are zygotically elmoPB[c06760]/Df(2R)Prl (A) or elmo8C6/Df(2R)Prl (B) exhibit a dramatic loss of muscles and increase in unfused myoblasts (A′, B′, arrows). (C-H) Wild-type and elmoPB[c06760]/Df(2R)Prl maternal/zygotic mutant embryos stained with anti-MEF2 (C, D) anti-Krüppel (Kr) (E, F), and anti-SNS (G, H) antibodies. (C,D) Similar number of MEF2-expressing cells are present in stage 14 wild-type and elmo mutants. (E, F) Stage 13 wild-type and elmo mutants exhibit founder cell positive cells as stained with the founder cell antibody Kr. (G, H) Similar staining patterns are observed with SNS antibody showing fusion competent cell fate is preserved in stage 13 wild-type and elmo mutants. All views are lateral, with anterior to the left and dorsal to the top. Scale bar, 50μM
Given the almost ubiquitous expression of elmo, and its requirement prior to myoblast fusion, we sought to determine whether the myoblast fusion defects observed in elmo mutants reflect an earlier role in muscle cell fate commitment. To address this possibility, we analyzed elmo maternal and zygotic mutants with various mesodermal markers. DMEF2 is expressed in all developing muscle cells and is required for their differentiation. Similar numbers of DMEF2 positive cells are observed in both WT and elmo mutant embryos (Fig. 6C, D), suggesting that a decrease in elmo does not impact their specification. Likewise, specification of founder cells and fusion competent cells appears to have occurred normally in elmo mutants, as visualized using antibodies against Kruppel (Kr) and Sticks and stones (SNS), respectively (Fig. 6E–H). To determine if the muscle defects observed in elmo mutants are autonomous to the muscle and not a result of secondary defects in other tissues, we expressed UAS-elmoIR in the muscle under the control of mef2-GAL4. Due to the presence of maternal load and the variability in which RNAi works in tissues, it is not surprising that gross defects in the final muscle pattern were not observed. However, an approximately two-fold increase in unfused myoblasts were observed in stage 16 embryos expressing two copies of elmo RNAi compared to mef2-GAL4 control embryos at 29°C. Presumably due to higher temperatures which can result in minor developmental defects, control embryos showed an average of 26.8 unfused myoblasts (SD=9.7; n=11) in hemisegments A1–A5 as visualized by immunofluorescence with anti-MHC (data not shown). Embryos expressing elmo RNAi in the musculature showed an average of 49.3 unfused myoblasts/A1–A5 hemisegments with a SD=9.0 (n=9) with a confidence level of >99.0%. This data strongly suggests that the elmo-associated defects in myoblast fusion are autonomous to the musculature, and due to a block in fusion rather than a decrease in properly specified cells.
MBC and ELMO function in concert to disrupt myoblast fusion
Mesodermal expression of constitutively-active Rac1 has previously been shown to have a negative impact on muscle development, severely impairing myoblast fusion (Luo et al., 1994). Since MBC and ELMO are predicted to function together to activate Rac1, by analogy to their orthologs, it was of interest to determine whether excess MBC and ELMO collaborate to perturb myoblast fusion. The mef2-GAL4 driver was used to direct expression of UAS-mbc, UAS-elmo, or both, to the developing musculature. Representative embryos from this analysis are shown in Figure 7, and demonstrate that neither an increase in MBC nor an increase in ELMO cause obvious muscle perturbations (Fig. 7A–C). By contrast, an excess of both MBC and ELMO has a profound effect on myoblast fusion (Fig. 7D), indicating that they work together to induce downstream events. These data are consistent with the results of our mass spectrometry analysis, which demonstrate that MBC and ELMO interact stoichiometrically in the musculature, and suggest that this is the functional form of the two proteins.
Figure 7.
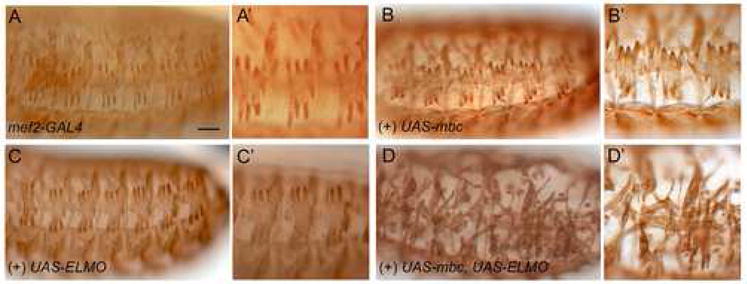
Overexpression analysis of CDM pathway members in the musculature. (A–D′) Low magnification (A, B, C, D) and high magnification (A′ B′, C′, D′) views of stage 16 embryos expressing UAS-cDNAs under control of the pan-mesodermal mef2-GAL4 driver. (A, A′) Embryos of the genotype mef2-GAL4 show a mature muscle pattern with no missing muscles and unfused myoblasts. (B–C′) Expression of UAS-mbc or UAS-elmo, respectively, has no effect on the muscle pattern. (D, D′) Co-expression of UAS-elmo and UAS-mbc results in missing muscles and many unfused myoblasts. All views are lateral, with anterior to the left and dorsal to the top. Scale bar, 50μM
MBC and ELMO function as a Rac-GEF in the developing eye
The Drosophila eye, where genetic interactions are easily observed, has been used extensively to study proteins required for GTPase signaling, including Rac1 and Rac2 (Hakeda-Suzuki et al., 2002; Nolan et al., 1998). Of note, mbc was identified as a suppressor of a rough eye phenotype induced by excess wild-type Rac1 (Nolan et al., 1998). To test the idea that MBC and ELMO act in concert to activate Rac in vivo, we analyzed genetic interactions in the eye. As in the embryonic muscle, neither excess MBC nor excess ELMO alone have an impact on ommatidial organization (Fig. 8A, B). However, coincident expression of MBC and ELMO causes a rough eye phenotype (Fig. 8C). The rough eye phenotype is suppressed by a reduction in endogenous mbc or elmo (Fig. 8E), confirming that these proteins function stoichiometrically and that the rough eye phenotype is not due to forced interactions with other proteins. More importantly, the rough eye phenotype appears to reflect activation of endogenous Rac proteins by the MBC/ELMO complex, since it is sensitive to the dosage of these GTPases (Fig. 8D).
Figure 8.
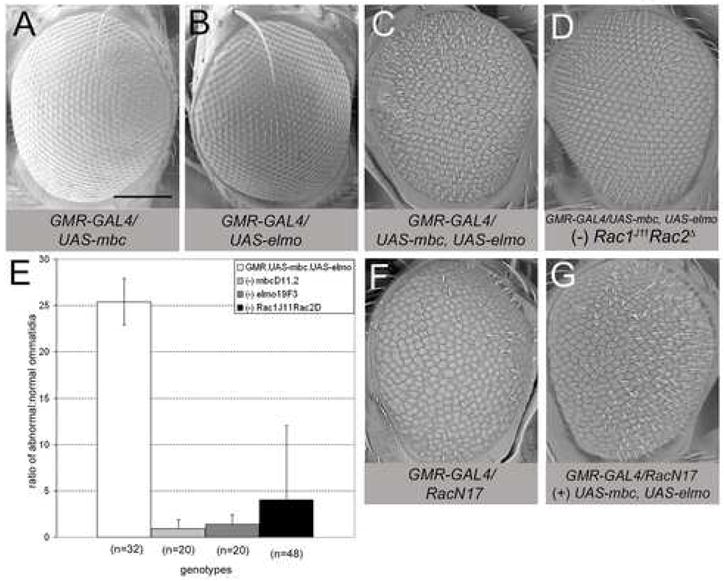
Genetic interactions between mbc, elmo, and Rac alleles. (A–D, F, G) Scanning electron micrographs of adult eyes expressing UAS-cDNAs under control of GMR-GAL4. (A, B) Overexpression of mbc or elmo has no apparent impact on eye morphology. (C) Co-expression of mbc and elmo results in a severe rough eye phenotype. (D) Removal of one copy of Rac1J11Rac2Δ suppresses the rough eye phenotype, resulting in nearly wild-type eye morphology. (E) Graph depicting the ratio of abnormal:normal (A:N) ommatidia present in flies co-expressing mbc and elmo compared to flies co-expressing mbc and elmo in a heterozygous background for mbc, elmo, or Rac1Rac2. Adult eyes co-expressing mbc and elmo (white bar) exhibit a ratio of 25:1 (n=32) while the ratio of abnormal:normal ommatidia decreases when heterozygous for mbcD11.2 (A:N=0.65; n=20), elmo19F3 (A:N=0.51; n=20) or Rac1J11Rac2Δ (A:N=1.32; n=48). n is the number of eyes examined for each genotype. See also Supp Fig. 2(F, G) Expression of RacN17 results in a severe rough eye morphology (F) that is suppressed by coincident expression of mbc and elmo (G). Scale bar, 200μM
As a final confirmation that MBC and ELMO act together to recapitulate Rac-GEF activity, we performed an analysis similar to that described by Huelsmann (Huelsmann et al., 2006). We took advantage of the severe rough eye phenotype caused by expression of RacN17, which is characterized by fused ommatidia and missing bristles ((Fan and M., 1994); Fig. 8F, Supp. Fig. 2). This dominant-negative form of Rac is thought to sequester free GEFs in the cell. Thus an increase in the MBC/ELMO complex would provide additional GEF to activate Rac, thereby suppressing the eye perturbations cased by RacN17. Indeed, co-expression of MBC and ELMO suppresses the rough eye phenotype caused by RacN17 (Fig. 8G, Supp. Fig. 2), indicating that these proteins function together as a Rac-GEF.
DISCUSSION
The CDM signaling pathway in Drosophila includes the DOCK180/CED-5 ortholog MBC. Like conventional GEFs, DOCK180 and CED-5 contain highly conserved domains that can bind to monomeric GTPases and contribute to their activation (Meller et al., 2005). However, the conserved PH-domain that is normally present within conventional GEFs is actually provided by a separate protein family represented by ELMO/CED-12 (Brugnera et al., 2002; Lu et al., 2004).. Thus, one member of each of these two protein families can combine to form an unconventional bipartite GEF. While the small adaptor protein Crk often forms a critical component of this complex, recent studies have suggested that both DOCK180 and MBC can function in its absence (Balagopalan et al., 2006; Tosello-Trampont et al., 2007). Moreover, Crk has been shown to function in pathways that are totally independent of this bipartite GEF (Sasahara et al., 2002; Tang et al., 2005). Reminscent of this diversity of interactions, recent studies have suggested that CDM and ELMO/CED-12 family proteins may also function through independent interactions. For example, DOCK180 is capable of activating Rac on its own and has positive effects on cell migration and phagocytosis, albeit enhanced by binding of ELMO (Brugnera et al., 2002; Katoh and Negishi, 2003; Lu et al., 2004), and ELMO interacts with radizin independent of its interaction with DOCK180 (Grimsley et al., 2005). Drosophila MBC functions in a wide variety of processes that include border cell migration in the ovary and myoblast fusion in the embryo (Duchek et al., 2001; Erickson et al., 1997) and can contribute to the activation of Rac1 in the eye (Nolan et al., 1998). We have demonstrated herein, through its biochemical and genetic analysis, that Drosophila elmo functions in concert with MBC in these processes. Like MBC, decreased levels of ELMO impair border cell migration and myoblast fusion ((Bianco et al., 2007) and studies herein). Moreover, MBC interacts stoichimetrically in the mesoderm with ELMO. Coincident over-expression of this complex impairs myoblast fusion, reinforcing the model from constitutively-active Rac1 that excess active Rac1 also interfers with myoblast fusion. Co-expression of MBC and ELMO also impacts development of the adult eye, resulting in a rough eye phenotype that is suppressed by decreasing endogenous levels of Rac. These data indicate that MBC and ELMO function together in a complex, and as a RacGEF.
ELMO is expressed and acts in concert with MBC during various stages in Drosophila development
As discussed above, CDM family proteins are required for a diverse array of biological processes. If ELMO functions with MBC in these processes, one would expect it to be expressed in a temporal and spatial pattern coincident with that of MBC. Consistent with this expectation, RT-PCR throughout the fly life cycle, embryonic in situ hybridizations and antibody stainings at multiple stages of Drosophila development, reveal that elmo is broadly expressed. Interestingly, however, the MBC/ELMO complex may serve distinct roles in each of the tissues in which it is expressed. MBC and ELMO are required for migration of the border cells in the ovary (Bianco et al., 2007; Duchek et al., 2001); however, myoblast migration appears to occur normally in mbc mutant embryos, as the fusion competent cells in mbc mutants can be found clustered and aligned with the founder cells
The CDM/ELMO(Ced-12) complexes in both vertebrate and C. elegans function upstream of Rac as unconventional bipartite GEFs to promote exchange of GDP for GTP in activation of monomeric GTPases (Meller et al., 2005). Our studies support a similar role for Drosophila MBC/ELMO. First, during ommatidial development, the rough eye phenotype resulting from co-expression of MBC and ELMO can be suppressed by removing half the gene dosage contributed by Rac1 and Rac2. Also, overexpression of the MBC/ELMO complex is sufficient to provide enough GEF activity to overcome the effect of its sequestration by RacN17 in the eye. In both the musculature and eye, expression of neither MBC nor ELMO alone has a phenotypic consequence, yet co-expression of MBC and ELMO phenocopies activated Rac.
Noteably, embryos that are completely lacking both maternal and zygotic elmo die before muscle development occurs, possibly reflecting an earlier role for the protein. Interestingly, however, the development of mutant embryos that lack both maternally and zygotically provided mbc continues until myogenesis (Balagopalan et al., 2006). These data suggest that, like its vertebrate counterparts, Drosophila ELMO has multiple binding partners. In addition to DOCK180, vertebrate ELMOs bind to three of the five additional CDM family members, suggesting that ELMO binding is a general feature of these proteins (Grimsley et al., 2004; Sanui et al., 2003). Based upon primary sequence homology, the fly genome contains at least four potential CDM superfamily members in addition to MBC (Cote and Vuori, 2002) The predicted transcripts of two of these are most closely related to vertebrate DOCK9/11 and DOCK 7/8. The Drosophila protein most closely related to vertebrate DOCK4 has been reported as the protein product of the sponge locus (Postner et al., 1992), but has not been studied extensively. It remains to be seen whether Drosophila ELMO is capable of binding to these other CDM-like molecules, and functions in concert with them in other tissues. One such place to examine in this regard is the embryonic CNS, where ELMO expression is quite strong but MBC is strikingly low (Erickson et al., 1997). Thus, alternative CDM/ELMO-like complexes may be present and required in different tissues throughout Drosophila development, or in the same tissues to regulate different GTPases.
Interaction of ELMO with other proteins and CDM/ELMO integration with other signaling pathways
The above studies, combined with recent reports of ELMO binding to non-CDM family members, may reflect a role for ELMO proteins in integrating signals from different pathways. Interaction of the N-terminal region of ELMO with RhoG is capable of translocating the ELMO/DOCK180 complex to the membrane to regulate neurite outgrowth and cell migration (Katoh and Negishi, 2003). Simultaneously, the N-terminal region of ELMO can bind to both the inactive and active forms of the ERM protein radixin (Grimsley et al., 2005). Interestingly, this ELMO/radixin interaction does not affect the ability of the ELMO/DOCK180 complex to promote Rac activation (Grimsley et al., 2005). Hence, ELMO may be functioning at the membrane to regulate the actin cytoskeleton via Rac, while recruiting radixin and ERM family members to perform their recognized roles in cross-linking the actin cytoskeleton to the plasma membrane.
In addition to Rac activation via the CDM/ELMO proteins, the ARF (ADP-ribosylaton factor) family of GTPases has been shown to signal through Rac (D’Souza-Schorey and Chavrier, 2006). Both DOCK180 and ELMO colocalize with ARNO (an ARF-GEF) and overexpression of mutant forms of DOCK180 and ELMO mutants block ARNO-induced Rac activation (Santy et al., 2005). However, RhoG signaling does not seem to be required for the ARNO-ELMO activation of Rac (Santy et al., 2005). This suggests Rac activation is differentially regulated in cell or tissue-specific manners or that within a cell there are localized mechanisms defined by crosstalk between signaling pathways that are responsible for Rac membrane localization. Intriguingly, in the Drosophila musculature, expression of a dominant-negative ARF6 results in myoblast fusion defects while the corresponding ARF-GEF Loner/Schizo is required for membrane localization of Rac (Chen et al., 2003). More work is required to see if ARF6, Loner/Schizo, and the ELMO/MBC proteins function in a signaling pathway in Drosophila to activate Rac.
Rac activity and myoblast fusion
Numerous studies have established a crucial role for Rac activation in myoblast fusion. More than a decade ago, Luo and colleagues demonstrated that a dominant-negative form of Rac1 interferes with myoblast fusion (Luo et al., 1994). A herculean genetic analysis of the small Rac GTPases proved that the fusion defects observed with dominant-negative Rac1 reflect the Rac1, Rac2 loss-of-function phenotype (Hakeda-Suzuki et al., 2002) The need for activated Rac in myoblast fusion was further supported by the phenotype of embryos mutant for mbc (Erickson et al., 1997) and elmo, as shown herein. Rac, in turn, likely functions to direct rearrgangement of the actin cytoskeleton through regulation of the Arp2/3 complex via Kette and WAVE (Chen and Olson, 2005). Surprisingly, however, constitutively-active forms of Rac negatively impact this process in a manner that, on the surface, have the same phenotypic consequence as the absence of active Rac (Luo et al., 1994). The specific relevance of excess activated Rac1 to normal myoblast fusion remains unclear, and we cannot rule out that excess Rac interferes with other signaling pathways that do not normally impact myoblast fusion. However, perturbation of this phenotype also has the potential to uncover key components and regulators that contribute to the normal process.
Our data establish, for example, that monomeric Rac GTPases are in excess, and are therefore available to be activated when the level of the MBC/ELMO GEF complex is increased. Under normal circumstances, then, the ability of the cell to activate this endogenous Rac remains low. Potential mechanisms for this include the direct regulation of MBC and/or ELMO levels through synthesis or turnover. Though little is known about the synthesis of either MBC or ELMO or their levels in the cell, it is intriguing that DOCK180 is ubiquitinated and this modification ensures its rapid turnover (Makino et al., 2006). Alternatively, GAPs may be present to ensure that Rac does not remain active. Finally, the MBC/ELMO pathway may be integrating with the loner-Arf6 associated pathway, as discussed above, in such a way that perturbation of MBC and ELMO is impacting Rac1 activity through components of the Arf6 pathway. Either way, the fact that this myoblast fusion phenotype is occurring in response to perturbation of the endogenous pathway by wild-type proteins in a stoichiometric manner suggests that it may be possible to modulate it in ways that provide insights into the myoblast fusion process.
Supplementary Material
Acknowledgments
We thank Jeff Haug and Josh Wunderlich of the Stowers Institute Cytometry and Molecular Biology Core Facilities for assistance. We thank members of the Abmayr lab, particularly Claude Shelton, Kiran Kocherlakota, David Ash and Maggie Chen for helpful discussions and assistance. ERG was supported by a Ruth Kirschstein NRSA fellowship; Grant Number F32A053027 from the National Institute of Arthritis and Musculoskeletal And Skin Diseases. This work was supported by the Stowers Institute for Medical Research and NIH grant RO1 AR44274 to S.M. Abmayr.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balagopalan L, et al. The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol Cell Biol. 2006;26:9442–55. doi: 10.1128/MCB.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, et al. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–5. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- Bourne HR. Rac and cell migration: CDM proteins integrate signals. Nat Cell Biol. 2005;7:777–8. doi: 10.1038/ncb0805-777. [DOI] [PubMed] [Google Scholar]
- Brugnera E, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4:574–82. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- Chang HY, Ready DF. Rescue of photoreceptor degeneration in rhodopsin-null Drosophila mutants by activated Rac1. Science. 2000;290:1978–80. doi: 10.1126/science.290.5498.1978. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev Cell. 2001;1:705–15. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–73. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Chen EH, et al. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–62. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- Cheresh DA, et al. Regulation of Cell Contraction and Membrane Ruffling by Distinct Signals in Migratory Cells. The Journal of Cell Biology. 1999;146:1107–1116. doi: 10.1083/jcb.146.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote JF, et al. A novel and evolutionarily conserved PtdIns(3,4,5)P(3)-binding domain is necessary for DOCK180 signalling. Nat Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J Cell Sci. 2002;115:4901–13. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- de Bakker CD, et al. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol. 2004;14:2208–16. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Duchek P, et al. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- Eng JK, et al. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Erickson MRS, et al. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure and cytoskeletal organization. Journal of Cell Biology. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C-MMT-L. Patterning of mammalian somites by surface ectoderm and notochord: evidence for schlerotome induction by a hedgehog homolog. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–75. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- Geisbrecht ER, Montell DJ. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 2004;118:111–25. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Grimsley CM, et al. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J Biol Chem. 2004;279:6087–97. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- Grimsley CM, et al. Characterization of a novel interaction between ELMO1 and ERM proteins. J Biol Chem. 2005;291:5928–5937. doi: 10.1074/jbc.M510647200. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, et al. Rac function and regulation during Drosophila development. Nature. 2002;416:438–42. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, et al. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Molecular and Cellular Biology. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann S, et al. The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development. 2006;133:2915–24. doi: 10.1242/dev.02449. [DOI] [PubMed] [Google Scholar]
- Ishimaru S, et al. PVR plays a critical role via JNK activation in thorax closure during Drosophila metamorphosis. Embo J. 2004;23:3984–94. doi: 10.1038/sj.emboj.7600417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461–4. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–9. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- Kiyokawa E, et al. Evidence that DOCK180 up-regulates signals from the CrkII-p130(Cas) complex. J Biol Chem. 1998;273:24479–84. doi: 10.1074/jbc.273.38.24479. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, et al. Membrane recruitment of DOCK180 by binding to PtdIns(3,4,5)P3. Biochem J. 2001;354:73–8. doi: 10.1042/0264-6021:3540073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, et al. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat Struct Mol Biol. 2004;11:756–62. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
- Lu M, et al. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr Biol. 2005;15:371–7. doi: 10.1016/j.cub.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Luo L, et al. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Makino Y, et al. Elmo1 inhibits ubiquitylation of Dock180. J Cell Sci. 2006;119:923–32. doi: 10.1242/jcs.02797. [DOI] [PubMed] [Google Scholar]
- McDonald WH, et al. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LCMS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom. 2002;219:245–251. [Google Scholar]
- Meller N, et al. CZH proteins: a new family of Rho-GEFs. J Cell Sci. 2005;118:4937–46. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- Niewiadomska P, et al. DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J Cell Biol. 1999;144:533–47. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KM, et al. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes Dev. 1998;12:3337–42. doi: 10.1101/gad.12.21.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti AC, et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–33. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postner MA, et al. Maternal effect mutations of the sponge locus affect actin cytoskeletal rearrangements in Drosophila melanogaster embryos. J Cell Biol. 1992;119:1205–18. doi: 10.1083/jcb.119.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif K, Cyster J. The CDM protein DOCK2 in lymphocyte migration. Trends Cell Biol. 2002;12:368–73. doi: 10.1016/s0962-8924(02)02330-9. [DOI] [PubMed] [Google Scholar]
- Santy LC, et al. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol. 2005;15:1749–54. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- Sanui T, et al. DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood. 2003;102:2948–50. doi: 10.1182/blood-2003-01-0173. [DOI] [PubMed] [Google Scholar]
- Sasahara Y, et al. Mechanism of recruitment of WASP to the immunological synapse and of its activation following TCR ligation. Mol Cell. 2002;10:1269–81. doi: 10.1016/s1097-2765(02)00728-1. [DOI] [PubMed] [Google Scholar]
- Tabb DL, et al. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang DD, et al. The adapter protein CrkII regulates neuronal Wiskott-Aldrich syndrome protein, actin polymerization, and tension development during contractile stimulation of smooth muscle. J Biol Chem. 2005;280:23380–9. doi: 10.1074/jbc.M413390200. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tosello-Trampont AC, et al. Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells. Cell Death Differ. 2007;14:963–72. doi: 10.1038/sj.cdd.4402094. [DOI] [PubMed] [Google Scholar]
- Washburn MP, et al. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 1998a;93:951–60. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998b;392:501–4. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- Wu YC, et al. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- Zhou Z, et al. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev Cell. 2001;1:477–89. doi: 10.1016/s1534-5807(01)00058-2. [DOI] [PubMed] [Google Scholar]
- Zybailov B, et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–47. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


