Figure 3.
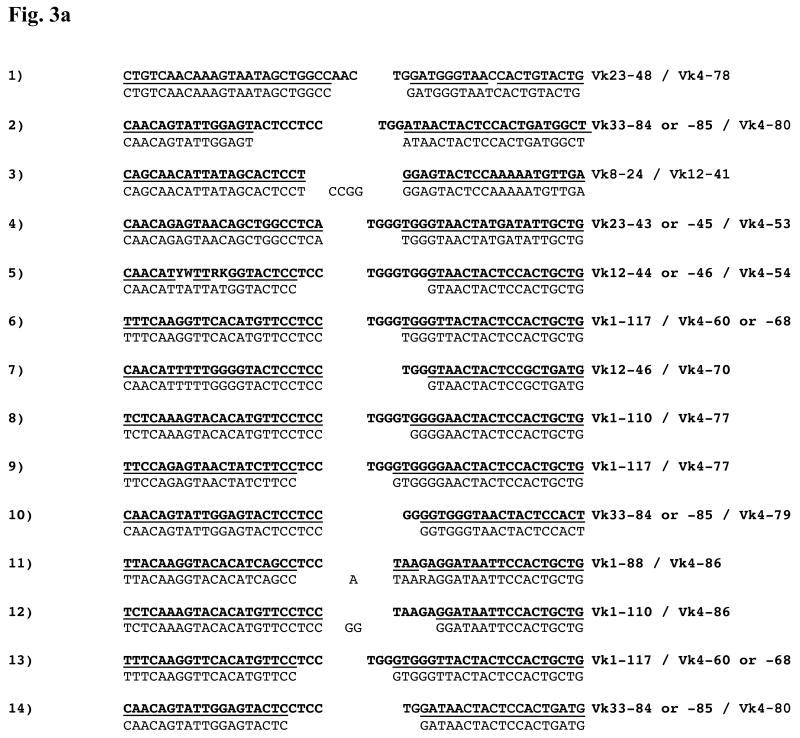
Figure 3a: Nucleotide Sequences of Vκ–Vκ junctions. Each Vκ gene contributes nucleotides from its 3′ end to the Vκ–Vκ junction. Here, the corresponding germline sequences appear in bold font above each junction to permit analysis of junctional modification. A bar between the germline and experimental sequences indicates regions of identity. Nucleotides that cannot be attributed to a particular germline sequence are shown centered in the junction. Displayed sequences are from the CDR3 of each gene, aligned against each other, and the Vκ families contributing to each junction are shown. The sequence numbering corresponds to the numbering in table 1, which provides the mouse/tissue/hybridoma origin of each sequence. For consistency, the top strands of the non-Vκ4 genes are shown on the left and the bottom strands of the Vκ4 genes (when present) are shown on the right. An example of one of these rearrangements (involving Vκ1-88 and Vκ4-86) and its corresponding signal joint is shown in fig. 2c.
Figure 3b: Sequences of other atypical κ rearrangements. Sequence data from the atypical junctions shown in fig. 4a-4c appear with the corresponding portions of the germline Igκ locus. The notation is as described for fig. 3a. In the hybrid joint involving the Jκ4 RSS, “<23bp>” indicates the spacer between the heptamer (shown, with junctional modification) and the nonamer (shown, preserved). Similarly, in the Vκ-Vκ signal joint, the 12-RSS spacer is denoted “<12bp>”. The DNA sources of the sequences are: 1. B6 spleen; 2. B6.bcl-xL spleen; 3. B6.bcl-xL spleen; 4. B6.56R.bcl-xL hybridoma; 5. B6.56R hybridoma; 6. B6.56R hybridoma.