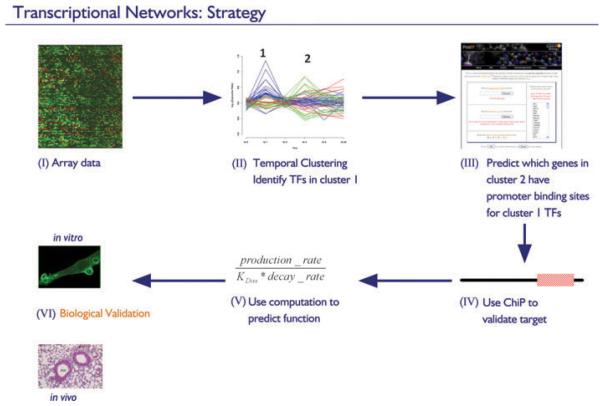
Fig. 4. Strategy for unraveling transcriptional networks.
(I) Microarrays are used to profile temporal gene expression responses in stimulated macrophages. (II) Clustering reveals groups of genes with different expression kinetics. (III) Candidate regulatory links between genes are made by searching for enrichment of regulatory elements of transcription factors expressed in early groups (no. 1) in the promoters of genes expressed in later groups (no. 2). (IV) Regulatory predictions are validated by chromatin immunoprecipitation. (V) Mathematical modeling is used to predict the functional nature of validated interactions (activation, repression, etc.). (VI) Functional predictions are tested in vitro and in vivo.