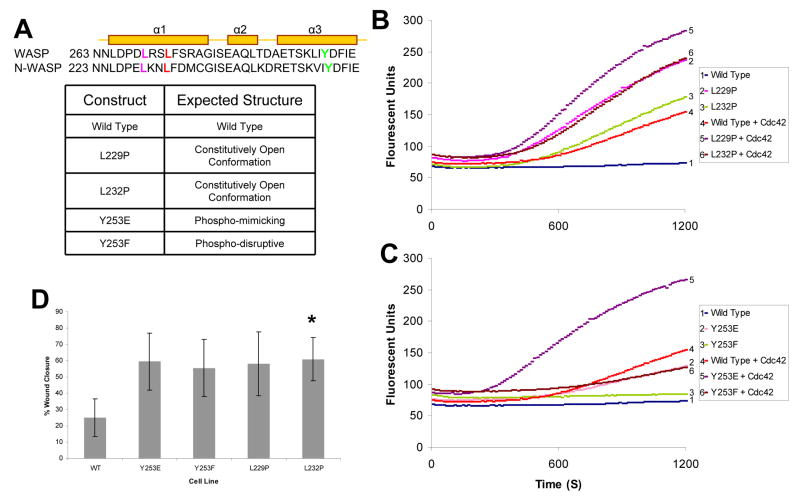
Fig. 1. NWASP mutations result in constitutive activation.
(A) Protein sequence alignment of a portion of the GBD of murine WASP and rat NWASP. Predicted tertiary structure of the sequence denoting α-helixes is shown [4]. The residues selected for mutations are L229 in purple, L232 in red, and Y253 in green. Table represents the predicted structural phenotypes for each of the NWASP mutants. (B) Pyrene actin assay measuring the actin polymerization activity of NWASP mutations predicted to have an open conformation in the presence/absence of Cdc42-GTPγS. (C) Pyrene actin assay with phospho-regulatory NWASP in the presence/absence of Cdc42-GTPγS. (D). Histogram demonstrating that NWASP-deficient fibroblasts expressing NWASP phospho-regulatory (Y253E and Y253F) or constitutively active (L229P & L232P) constructs result in increased migration in a scratch assay (* p< 0.05).