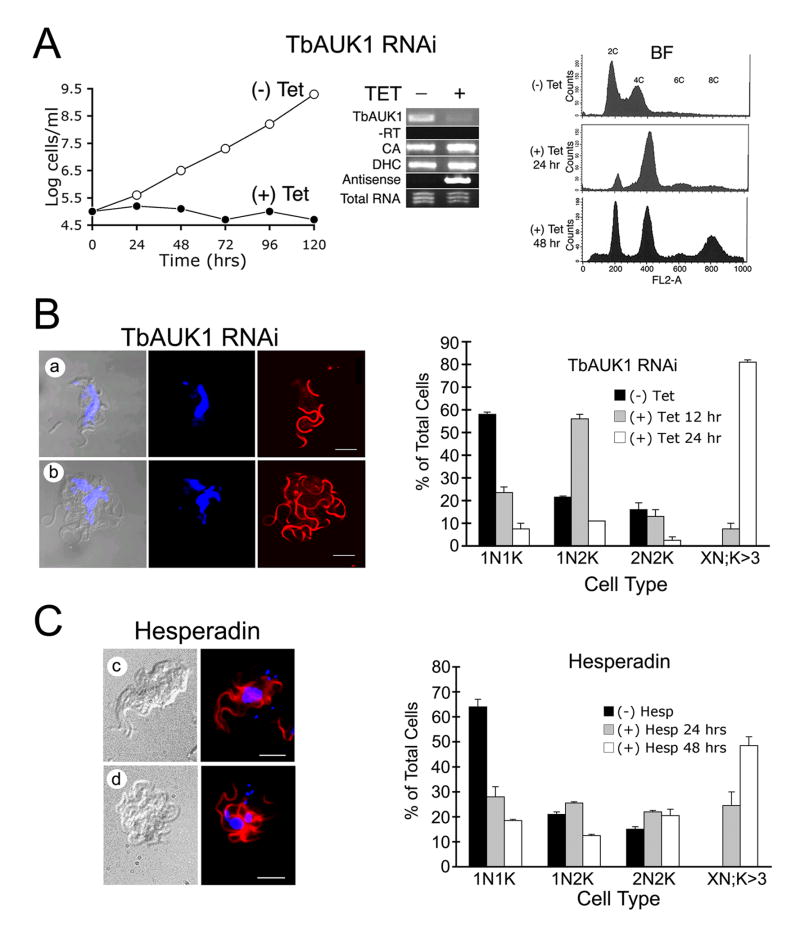
Fig. 6.
Treatment with Hesperadin phenocopies RNAi knockdown of TbAUK1.
(A) RNAi of TbAUK1 disrupts cell cycle progression in BF. The left panel shows a growth curve of TbAUK1 RNAi cells diluted to 1×105 cells/ml with (+) or without (-) tetracycline. The middle panel is RT-PCR using template RNA isolated from the TbAUK RNAi BF cells after growth for 72 hours with or without tetracycline. Primer pairs were designed to amplify the following: TbAUK1; the upstream gene carbonic anhydrase (CA); the downstream gene dynein heavy chain (DHC); and antisense TbAUK1. Also shown is the PCR control without reverse transcriptase (-RT) and the loading control of ethidium bromide stained total RNA (1 μg). The right panel shows flow cytometry of BF cells at increasing times after RNAi induction. A shift in population to 4C and 8C DNA content is observed.
(B) Changes in cell cycle progression induced by RNAi of TbAUK1. The left panels show morphological changes in BF after 24 hr induction with tetracycline. Cells were viewed by DIC, or stained for nuclei (TOTO) and flagella (PFR) (panels a-b). The right panel shows cell cycle progression in the RNAi cells. BF cultures were untreated, or induced with tetracycline for the amount of time indicated. After DAPI staining, cells were evaluated microscopically for their number of nuclei (N) and kinetoplasts (K). Cells designated 1N1K have one nucleus and one kinetoplast. Each time point is the evaluation of at least 200 cells (n=2, ±SE).
(C) Changes in cell cycle progression induced by Hesperadin. The left panels show morphological changes in cells treated with 100 nM Hesperadin for 24 hr (panels c-d); The right panel shows cell cycle progression of Hesperadin treated cells. The methods are the same as described in panel B.