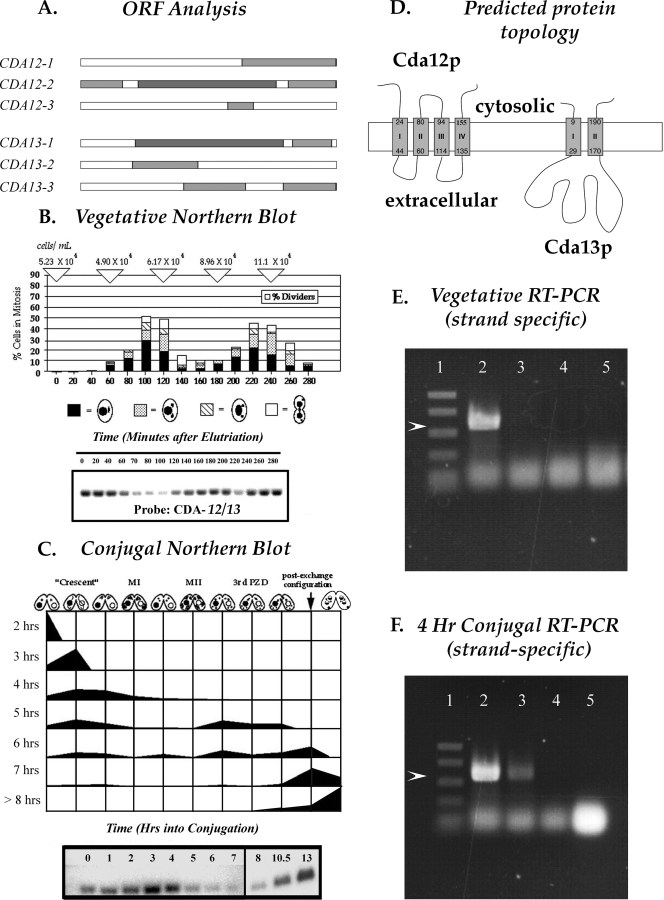
FIG. 1.
Analysis of transcripts from the CDA12/CDA13 gene locus. (A) ORFs at the CDA12/CDA13 locus (predicted by the NCBI ORF finder program). The diagram shows complete nesting of the smaller CDA12 ORF (CDA12-2) within the larger CDA13 ORF (CDA13-1). (B) Northern blot analysis using a double-stranded probe, showing transcription from the CDA12/CDA13 locus at various stages of cell division. Transcript levels drop as cells enter mitosis. (C) Northern blot analysis using a double-stranded probe, showing transcription from the CDA12/CDA13 locus at various stages during conjugation. Transcript levels rise during meiosis (3 to 4 h into conjugation) and again late in conjugation as pairs prepare to separate. MI, meiosis I; MII, meiosis II; 3rd PZD, third prezygotic division. (D) Protein topology predicted for CDA12 and CDA13 products (TopPred software prediction). Arabic numerals indicate amino acid positions. (E) Strand-specific RT-PCR analysis revealing that only CDA12 transcripts (lane 2) are present during vegetative growth. (Lane 1) PCR size markers (Promega; 1 kb and 750, 500, 300, and 150 bp). (Lane 2) RT-PCR product obtained using vegetative RNA, a unidirectional primer complementary to the CDA12 transcript, and RT. The PCR mixture was subjected to RNase treatment and heat inactivation of the RT enzyme, and then both CDA12 and CDA13 primers and Taq polymerase were added. The visible product is indicated by the arrowhead. (Lane 3) The same procedure described in the legend to lane 2 was performed, with the CDA12 unidirectional primer replaced with one complementary to the CDA13 transcript (no product is visible). (Lane 4) Control lane in which both primers were added only after RNase treatment and heat inactivation (demonstrates the effectiveness of RNase and heat treatment in terminating RT activity). (Lane 5) Both primers were added at the start but with only Taq polymerase, no RT, present (demonstrating the lack of PCR activity from Taq polymerase operating off of either RNA or contaminating DNA templates). Strand-specific primers were Lumpy 13 and Lumpy 14 (Table 1). (F) Results of an RT-PCR experiment similar to that described in the legend to panel E, except using RNA from conjugating cells at the 4-h time point rather than RNA from vegetative cells. Products from both CDA12 and CDA13 transcripts (arrowhead; lanes 2 and 3, respectively) are detectable. Lanes correspond to those in panel E.