Abstract
Bacteria are able to cope with the challenges of a sudden increase in salinity by activating adaptation mechanisms. In this study, exponentially growing cells of the pathogen Bacillus cereus ATCC 14579 were exposed to both mild (2.5% [wt/vol] NaCl) and severe (5% [wt/vol] NaCl) salt stress conditions. B. cereus continued to grow at a slightly reduced growth rate when it was shifted to mild salt stress conditions. Exposure to severe salt stress resulted in a lag period, and after 60 min growth had resumed, with cells displaying a filamentous morphology. Whole-genome expression analyses of cells exposed to 2.5% salt stress revealed that the expression of these cells overlapped with the expression of cells exposed to 5% salt stress, suggesting that the corresponding genes were involved in a general salt stress response. Upregulation of osmoprotectant, Na+/H+, and di- and tripeptide transporters and activation of an oxidative stress response were noticeable aspects of the general salt stress transcriptome response. Activation of this response may confer cross-protection against other stresses, and indeed, increased resistance to heat and hydrogen peroxide could be demonstrated after preexposure to salt. A temporal shift between the transcriptome response and several phenotypic responses of severely salt-stressed cells was observed. After resumption of growth, these cells showed cellular filamentation, reduced chemotaxis, increased catalase activity, and optimal oxidative stress resistance, which corresponded to the transcriptome response displayed in the initial lag period. The linkage of transcriptomes and phenotypic characteristics can contribute to a better understanding of cellular stress adaptation strategies and possible cross-protection mechanisms.
Bacillus cereus is a spore-forming gram-positive bacterium that is frequently isolated from foods (6, 43). It is able to cause two types of gastrointestinal diseases, emesis and diarrhea. The emetic syndrome (an intoxication) is caused by a heat-, acid-, and trypsin-stable toxin, cereulide, and the symptoms may occur after ingestion of foods contaminated with the toxin (9, 34). The diarrheal syndrome (a toxicoinfection) is caused by one or more enterotoxins produced by vegetative cells of B. cereus in the small intestine. The number of B. cereus cells required to cause disease is relatively high, and in most cases at least 105 CFU g−1 food has been found in foods implicated in disease (34).
Salt is widely used as a food additive in the food industry to control bacterial growth (1). Bacteria employ several strategies to adapt to adverse conditions, and upon activation of the so-called adaptive stress response bacteria can become more robust. Exposure to sodium chloride has been shown to induce a protective response in B. cereus, which enables this organism to survive under otherwise lethal conditions (5, 8, 26). The use of various mild preservation methods in foods is becoming more prevalent, underlining the significance of a better understanding of the stress responses of pathogens to ensure the microbial safety of foods.
When bacteria are challenged by increased salinity in their environment, water efflux occurs, and this results in a reduction in the cellular turgor. To restore and maintain cellular turgor, bacteria initiate a two-step adaptation response. Initially, K+ is taken up by the bacterial cell, and subsequently osmoprotectants, like glycine betaine, are imported by transport systems (21, 24, 30). For Bacillus subtilis five osmoprotectant uptake transport systems (OpuA to OpuE) have been described (21, 24, 30). OpuA, OpuB, and OpuC are multicomponent ABC transporters, whereas the OpuD and OpuE symporters each consist of a single component. In the genome sequence of B. cereus ATCC 14579 (18), several open reading frames putatively encoding osmoprotectant transporters have been identified, indicating the importance of these transporters in B. cereus ATCC 14579. To date, limited information is available about the underlying mechanisms of the salt stress response of B. cereus. A proteome study performed by Browne and Dowds (5) showed that enzymes involved in the central metabolism were induced in B. cereus NCIMB 11796 during exposure to salt stress. Furthermore, salt stress is known to induce the alternative transcription factor σB protein in B. cereus ATCC 14579 (35), suggesting a role for this regulator in the salt stress response of this strain.
In this study, we performed genome-wide comparative transcriptional analyses of B. cereus ATCC 14579 in response to 2.5% and 5% sodium chloride and combined these analyses with phenotypic analyses. The transcriptome profiles for the two salt stress conditions were compared in order to investigate the overlap in the responses, the so-called general salt stress response, and to identify specific responses of mildly and severely salt-stressed cells. Moreover, we linked observed transcriptome expression patterns to several responses of the salt-stressed cells.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
Mesophilic B. cereus strain ATCC 14579 was used throughout this study. The culture used was stored frozen in brain heart infusion (BHI) broth (Becton Dickinson, France) supplemented with 25% (vol/vol) glycerol (Sigma, The Netherlands) at −80°C. The bacteria were cultivated before each experiment in 10 ml BHI broth and incubated at 30°C with shaking at 200 rpm (Innova 4335; New Brunswick Scientific, The Netherlands) for 12 to 18 h. To produce two independent biological replicates of exponentially growing cultures, two stationary-phase cultures were diluted 1:100 (vol/vol) in two Erlenmeyer flasks (250 ml) containing 50 ml fresh BHI broth. The flasks were incubated at 30°C with shaking at 200 rpm until the cells were growing exponentially (absorbance at 600 nm, 0.4 to 0.5; Novaspec II spectrophotometer; Pharmacia Biotech, United Kingdom). When an optical density at 600 nm (OD600) of 0.4 to 0.5 was reached, decimal dilutions were prepared using 9 ml of a peptone saline solution (1 g neutralized bacteriological peptone [Oxoid, United Kingdom] supplemented with 8.5 g NaCl per liter) to determine the viable counts at time zero. Fifty-microliter aliquots of the appropriate dilutions were surface plated on BHI agar plates (BHI broth supplemented with 15 g agar [Oxoid] per liter) using a spiral plater (Eddy Jet; IUL Instruments, Spain). To study the effects of exposure to mild and severe salt stress on the growth of B. cereus ATCC 14579, BHI broth supplemented with 7.5% and 15% (wt/vol) NaCl, respectively, was added (the final supplementary concentrations of NaCl were 2.5%, and 5% [wt/vol], respectively), after which the cultures were incubated further at 30°C with shaking at 200 rpm. At constant intervals, appropriately diluted aliquots of the mildly and severely salt-stressed cells were plated on BHI agar plates. The plates were subsequently incubated for 16 to 24 h at 30°C, and the results were expressed in log10 CFU ml−1.
RNA isolation.
RNA was isolated from both unstressed and salt-stressed cultures. To do this, four 50 ml-cultures were incubated in 250 ml-Erlenmeyer flasks at 30°C with shaking at 200 rpm. When the absorbance at 600 nm reached 0.4 to 0.5, one culture was used for extraction of RNA for the reference unstressed sample (time zero). Twenty milliliters of this unstressed culture was transferred into a 50-ml Falcon tube (Greiner Bio-One, Germany) and processed as described below. The remaining three cultures were exposed to 2.5% NaCl by adding 25 ml of BHI broth supplemented with 7.5% NaCl and incubated further at 30°C with shaking at 200 rpm. At three time points, after 10, 30, and 60 min of exposure to salt, 30 ml of the mildly salt-stressed cells was transferred into 50-ml Falcon tubes. A similar procedure was used to expose the cells to 5% NaCl for 10, 30, and 60 min. Subsequently, the unstressed and salt-stressed cultures were pelleted by centrifugation at 15,000 × g at 4°C for 30 s (Eppendorf 5804 R centrifuge; Eppendorf, Germany). After the supernatant was decanted, 1 ml of phenol-based RNA extraction buffer (TRI reagent; Ambion, United Kingdom) was added to the cell pellets. Subsequently, the cell pellets were resuspended, snap-frozen in liquid nitrogen, and stored at −80°C. The experiments used to obtain RNA samples were performed in duplicate. After the cell pellets were defrosted, the cells were disrupted by bead beating (Mini Beadbeater-8; Biospec Products, United States) with zirconium beads (diameter, 0.1 mm; Biospec Products), and the RNA was isolated as previously described (35).
cDNA synthesis, Cy dye labeling, and microarray hybridization.
cDNA synthesis and Cy3 and Cy5 dye labeling of the cDNA samples were performed as previously described (37). Subsequently, the Cy5-labeled cDNA samples prepared from the mildly and severely salt-stressed cells were hybridized (concentration ratio, 1:1; 300 to 400 ng Cy dye-labeled cDNA per sample) with the Cy3-labeled reference sample (time zero). Independent biological duplicates of the reference sample and the mildly and severely salt-stressed samples were hybridized with the dyes swapped. After hybridization in a hybridization oven (Agilent), the microarrays were washed and blown dry with nitrogen gas.
In this study, custom-made B. cereus ATCC 14579 microarrays were used, which were printed by Agilent.
Microarray scanning and data analysis.
The microarrays were scanned using an Agilent microarray scanner (G2565BA). The data were extracted from the microarrays using Agilent's Feature Extraction software (version 8.1.1.1). The data extraction procedure included LOWESS normalization for the raw data. The web-based VAMPIRE microarray suite (17) was used for further statistical analysis of the normalized data. The data for duplicate experiments were merged, and significantly differentially expressed microarray spots were identified using a threshold for the false discovery rate of less than 0.05. Subsequently, the expression ratios of the spots representing the same open reading frame were averaged. An open reading frame was considered to be differentially expressed when all spots representing the open reading frame were significantly differentially expressed based on the VAMPIRE output. The transcriptional expression data for a total of 5,058 open reading frames for 2.5% and 5% salt-stressed cells were compared.
Catalase activity absorbance assay.
The catalase activity absorbance assay used was based on a previously described experimental procedure (7, 38). Untreated exponentially growing cells and cells which were salt stressed (2.5% or 5% NaCl) for 10 to 60 min were washed in phosphate-buffered saline (PBS) (138 mM NaCl, 2.7 mM KCl, 140 mM Na2HPO4, 1.8 mM KH2PO4; pH adjusted to 7.4 with HCl) and subsequently diluted (1:10, vol/vol) in PBS supplemented with H2O2 (final concentration, 40 mM). The decrease in absorbance at 240 nm was measured over time at 30°C with a spectrophotometer (Spectramax Plus 384; Molecular Devices, United States). One unit of catalase activity was defined as a decrease in the absorbance at 240 nm of 1 unit per minute. The rate of decrease for each sample was corrected for the amount of untreated or salt-stressed cells added to the assay buffer (absorbance at 600 nm and the assay dilution factor, as cell suspensions were diluted 1:10 when they were exposed to H2O2). For all experimental conditions, three replicates were performed.
Fluorescence microscopy.
Exponentially growing cultures were exposed to 5% NaCl for 150 min and subsequently double stained; 30 μM of the red fluorescent membrane dye FM 4-64 (Invitrogen, The Netherlands) and 2 μM of the green fluorescent nucleic acid dye SYTO-9 (Invitrogen) were added to 1-ml aliquots of a culture and incubated for 15 min at room temperature in the dark. Cells were imaged using a Zeiss Axioskop fluorescence microscope with a fluorescein isothiocyanate filter set (magnification, ×1,000; Carl Zeiss, Germany). Images were obtained with a Canon Powershot G3 digital camera.
Chemotaxis assays.
Exponentially growing cultures and cultures which were exposed to 5% NaCl for 10, 60, and 150 min were pelleted by centrifugation at 15,000 × g at 4°C for 1 min (Eppendorf centrifuge 5804 R; Eppendorf). The sample volume of the salt-stressed cells was adjusted based on the equivalent cellular density of unstressed cells (OD600) in 50 ml. The pellets of unstressed and salt-stressed cells were resuspended in 30 ml of PBS agar (PBS supplemented with 0.3% [wt/vol] agar) at 42°C, resulting in a cellular density in the agar of approximately 8 log10 CFU ml−1 agar. Each mixture was equally divided and poured in four 9-cm-diameter petri dishes. Sterile filter disks (6-mm blank paper disks; Becton Dickinson) were placed in the middle of the petri dishes on the solidified cell-agar mixture, and subsequently, 5-μl aliquots of stock solutions of trehalose and alanine (0.4 M) were pipetted on top of the disks. The plates were incubated at 30°C, and after 1, 2, 3, and 4 h of incubation the plates were examined for chemotaxis.
Determination of heat resistance and H2O2 resistance of salt-stressed cells.
Exponentially growing cultures and cultures which were exposed to 2.5% and 5% NaCl for 10 to 150 min were added to 20 ml preheated BHI broth (1:100, vol/vol) at 50°C and then incubated at 50°C with shaking at 200 rpm (Julabo SW22; Julabo Labortechnik, Germany). Before exposure and after 1, 2, 4, 6, and 8 min of exposure to 50°C, samples were taken, and appropriate dilutions were surface plated in duplicate on BHI agar plates. The plates were incubated for 16 to 24 h at 30°C. A similar procedure was used to determine the H2O2 resistance of exponentially growing cultures and salt-stressed cultures exposed to 1 mM H2O2. Both the heat and H2O2 inactivation experiments were performed in duplicate.
Microarray data accession number.
The microarray data have been deposited in the GEO database under accession number GSE13713.
RESULTS
Effect of 2.5% and 5% NaCl stress on growth kinetics.
To investigate the effect of exposure to mild and severe salt stress on the growth kinetics of B. cereus ATCC 14579, exponentially growing cultures (OD600, ∼0.4 to 0.5) were challenged with 2.5% and 5% NaCl. Exposure to 2.5% salt resulted in a slightly reduced growth rate (Fig. 1). Cultures challenged with 5% salt showed an initial lag phase. After this lag phase, cellular growth resumed during exposure to 5% salt and resulted in an increase in absorbance (Fig. 1a) at 60 min. However, this resumption of growth did not result in an increase in the viable counts (Fig. 1b). Microscopic observations of the cells revealed that filamentous cells were formed during exposure to 5% salt for more than 60 min. This explains the discrepancy between the observed absorbance values and the viable counts after the resumption of growth during exposure to severe salt stress.
FIG. 1.
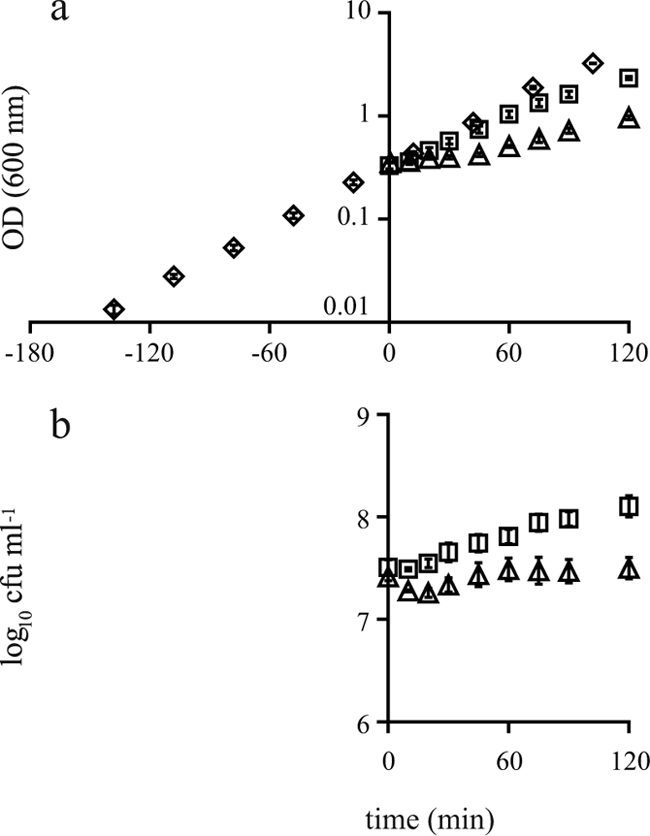
Growth (OD600 [a] and viable counts [b]) of B. cereus ATCC 14579 in BHI broth without addition of salt (⋄) and after addition of 2.5% salt (□) and 5% salt (▵) at time zero. The error bars indicate standard errors.
General transcriptome response to NaCl stress.
Microarray analyses were performed to obtain insight into the molecular response of B. cereus ATCC 14579 to both mild and severe salt stress. Exponentially growing cells were exposed to 2.5% and 5% salt, and samples were taken after 10, 30, and 60 min of exposure and compared to an untreated reference sample. The number of differentially expressed genes for each condition (2.5% salt for 10 min, 2.5% salt for 30 min, 2.5% salt for 60 min, 5% salt for 10 min, 5% salt for 30 min, and 5% salt for 60 min) is shown in Fig. 2. This figure shows that the number of differentially expressed genes in 2.5% salt-stressed cells was lower than the number of differentially expressed genes in 5% salt-stressed cells. Exposure of cells to 2.5% salt resulted in 153 differentially expressed genes at 10 min. After this first sampling time point, the number of differentially expressed genes decreased for the 30- and 60-min time points, at which there were 45 and 63 differentially expressed genes, respectively.
FIG. 2.
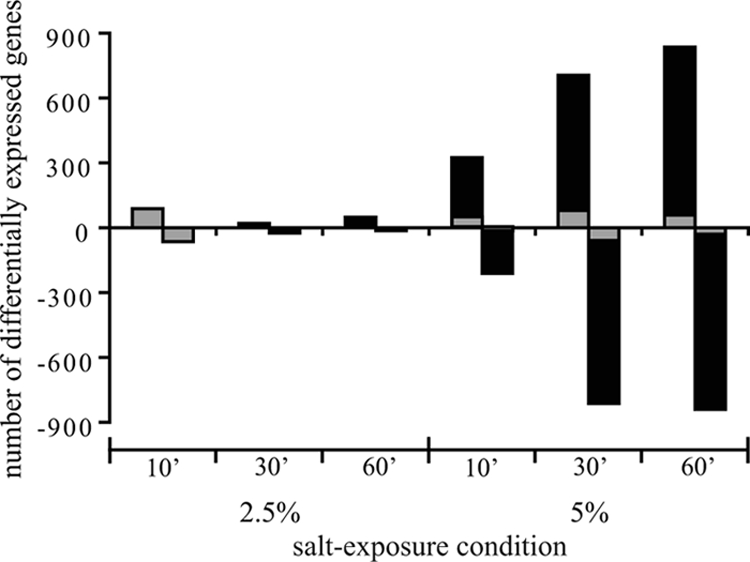
Numbers of differentially expressed genes in B. cereus ATCC 14579 after exposure to 2.5% and 5% salt for 10, 30, and 60 min. The gray bars indicate the genes that were differentially up- or downregulated after exposure to 2.5% salt for 10 min, as well as at the time points indicated.
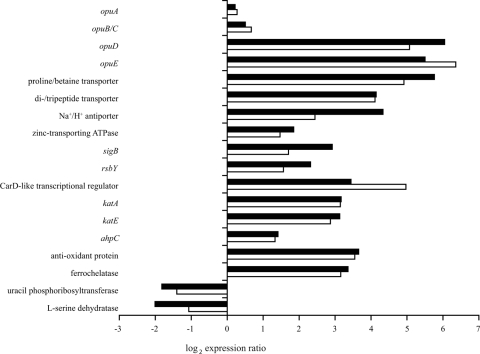
Approximately 95% of the differentially expressed genes (n = 147) in 2.5% salt-stressed cells exposed for 10 min were also differentially expressed at one or more time points during exposure to 5% salt. The overlapping transcriptome responses during mild and severe salt stresses included upregulation of genes encoding transporters involved in osmoregulation, genes encoding transcriptional regulators, and oxidative stress response-related genes and downregulation of genes involved in nucleotide and amino acid transport and metabolism. To provide more insight into the overlap of the transcriptome responses during exposure to mild and severe salt stresses, a selection of representative genes is shown in Fig. 3 (for a complete list of genes differentially expressed in cells exposed to 2.5% salt for 10 min and in cells exposed to 5% salt stress at one or more time points, see Table S1 in the supplemental material). To optimally visualize the overlap in gene expression, Fig. 3 shows the expression ratio after exposure to 2.5% salt for 10 min and the average ratio after exposure to 5% salt for 10, 30, and 60 min for each gene included. In the genome of B. cereus ATCC 14579 genes encoding various osmoprotectant transporters were identified (18). The osmoprotectant symporter genes opuD (BC0555) and opuE (BC3644), a gene encoding a proline/betaine transporter (BC3000) belonging to the major facilitator transporter family, and a gene encoding a proton-dependent di- and tripeptide transporter (BC0684) were among the most highly induced genes upon exposure to salt stress. In contrast, the genes encoding the multicomponent osmoprotectant ABC transporters OpuA (represented by BC2791) and OpuB/OpuC (represented by BC2232) were not differentially expressed during exposure to 2.5% and 5% salt stress. Furthermore, the upregulation of Na+/H+ antiporters (represented by BC1612) and other cation transporters (represented by a zinc-transporting ATPase gene, BC0596) was induced during both 2.5% and 5% salt stress. The transport of cations may contribute to the cation homeostasis inside the cell upon exposure to NaCl stress. These findings emphasized the importance of osmoprotectant, sodium, and other cation transporters in the salt stress response of B. cereus ATCC 14579.
FIG. 3.
Representative genes which were differentially expressed in B. cereus ATCC 14579 after exposure to both 2.5% and 5% salt. The filled bars indicate the expression ratios after exposure to 2.5% salt for 10 min, and the open bars indicate the average ratios after exposure to 5% salt for 10, 30, and 60 min. See Table S1 in the supplemental material for a complete list of the genes which were differentially up- or downregulated after exposure to 2.5% salt for 10 min and the expression ratios for these genes after 30 and 60 min of exposure to 2.5% salt and after 10, 30, and 60 min of exposure to 5% salt.
The expression of the gene encoding the alternative transcription factor σB (sigB, BC1004), which is involved in the global adaptation response to stress, was upregulated during exposure to both mild and severe salt stresses. Transcription of sigB in cells exposed to 5% salt increased over time, and the expression ratio at 60 min was comparable to that of cells exposed to 2.5% salt for 10 min. Likewise, the activator of σB, rsbY (BC1006), and many other members of the σB regulon (37) were also upregulated. This finding supported the putative involvement of σB in the salt stress response of B. cereus ATCC 14579. However, no differences in the growth kinetics were observed when exponentially growing cells of the sigB deletion mutant FM1400 (35), the rsbY deletion mutant FM1401 (36), and parental strain ATCC 14579 were exposed to 2.5% and 5% salt (data not shown). In addition to upregulation of the transcriptional regulator gene sigB, two putative CarD-like transcriptional regulators (25) were highly expressed (represented by BC4714).
Exposure to both mild and severe salt stresses induced genes that have been described to be involved in the oxidative stress response of bacterial cells. The main vegetative catalase gene katA (BC1155) and the σB-dependent catalase gene katE (BC0863) (37) were upregulated. In addition, an alkyl hydroperoxide reductase gene, ahpC (BC0377), that has been demonstrated to be involved in scavenging endogenous hydrogen peroxide in Escherichia coli (28), and a gene encoding an antioxidant protein (BC5044) were found to be upregulated. In cultures of B. subtilis grown under high-salinity conditions, various iron limitation-related genes were upregulated (13, 33). In this study we also observed induction of genes involved in iron homeostasis, including a gene encoding a ferrochelatase (BC1154), which is neighbor of katA.
The overlap in the responses during exposure to 2.5% and 5% salt not only encompassed upregulated genes but also included repression of 61 genes (see Table S1 in the supplemental material). Several genes involved in nucleotide transport and metabolism (represented by a uracil phosphoribosyltransferase gene, BC3891) and in amino acid transport and metabolism (represented by an l-serine dehydratase gene, BC4136) were repressed upon exposure to salt stress. It is likely that a reduced growth rate allowed reductions in amino acid and pyrimidine metabolism, and this is consistent with previous findings for B. subtilis (33) and E. coli (40).
Transcriptome response-specific 5% NaCl stress.
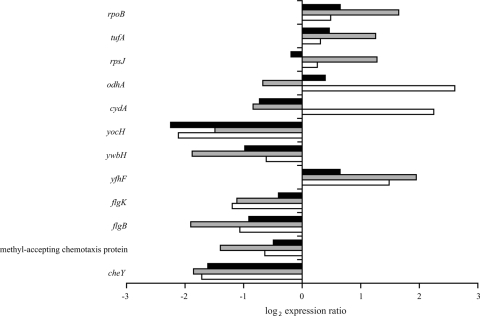
Exposure of B. cereus ATCC 14579 to 5% salt stress resulted in a greater transcriptome response than exposure to 2.5% salt stress (Fig. 2). The observed differences between the growth kinetics during exposure to severe salt stress and the growth kinetics during exposure to mild salt stress may have contributed to the 5% salt stress-specific transcriptome profile. Genes putatively involved in various biological processes were found to be up- or downregulated in cells exposed to severe salt stress. Figure 4 shows the expression ratios for genes which were representative of the 5% salt stress-specific transcriptome response. The expression ratios for the representative genes after exposure to 5% salt for 10 min, 30 min, and 60 min are shown in this figure.
FIG. 4.
Representative genes which were differentially expressed in B. cereus ATCC 14579 during exposure to 5% salt. The black bars, gray bars, and open bars indicate the expression ratios after exposure to 5% salt for 10, 30, and 60 min, respectively.
Exposure to 5% salt stress resulted in an initial lag phase before growth had resumed after 60 min of exposure. The transcriptome analysis revealed that genes involved in protein synthesis were upregulated during the lag phase at 30 min with exposure to 5% salt. Genes encoding an RNA polymerase (represented by rpoB [BC0122]), protein translation elongation factors (represented by tufA [BC0129]), and large- and small-subunit ribosomal proteins (represented by rpsJ [BC0130]) were induced. After 60 min of exposure upregulation of tricarboxylic acid cycle genes (represented by the 2-oxoglutamate dehydrogenase gene, odhA [BC1252]) was observed to correspond to the resumption of growth at this time point. Increased expression of tricarboxylic acid cycle proteins after resumption of growth was demonstrated previously for salt-challenged B. subtilis cells (15). Our transcriptome data showed that components of the electron transport chain were also induced at 60 min (represented by the cytochrome d ubiquinol oxidase subunit I gene, cydA [BC1938]).
Resumption of growth resulted in formation of filamentous cells. Autolysins are involved in hydrolyzing peptidoglycan, which allows the daughter cell to separate (31), and indeed, in our study the putative autolysin gene yocH (BC0679) and genes encoding a murein hydrolase exporter (represented by ywbH [BC3669]) were downregulated during 5% salt stress. Moreover, the putative cell division inhibitor yfhF (BC0497) gene was induced.
Exposure to severe salt stress affected the expression of genes involved in cell motility. Genes encoding proteins of the flagellar apparatus (represented by flgK [BC1636] and flgB [BC1641]) were repressed. Furthermore, genes described to be involved in chemotaxis were downregulated. For example, genes encoding methyl-accepting chemotaxis proteins, also known as chemoreceptors (represented by BC0422), and genes involved in the regulation cascade of chemotaxis (represented by cheY [BC1627]) were repressed.
Oxidative response is activated in NaCl-adapted cells.
Catalase activity is an important defense mechanism against hydrogen peroxide stress. Exposure to both 2.5% and 5% salt stresses resulted in induction of the expression of genes encoding catalases, represented by katA and katE in Fig. 3. Therefore, catalase activity in exponentially growing cells and in cells exposed to 2.5% and 5% salt stresses was assessed (Fig. 5). The catalase activity of 2.5% salt-adapted cells was significantly (P < 0.05) higher than that of unstressed cells. Five percent salt stress induced a lag phase, and cells in this adaptation phase (5% salt for 30 min) did not show increased catalase activity despite upregulation of kat genes. When cellular growth had resumed at 60 min, the catalase activity in the cells adapted to severe salt stress was significantly higher (P < 0.05) than that in unstressed cells. The increased catalase activity in cells adapted to both mild and severe salt stresses was confirmed by visualization of the catalase activity on a native polyacrylamide gel (data not shown).
FIG. 5.
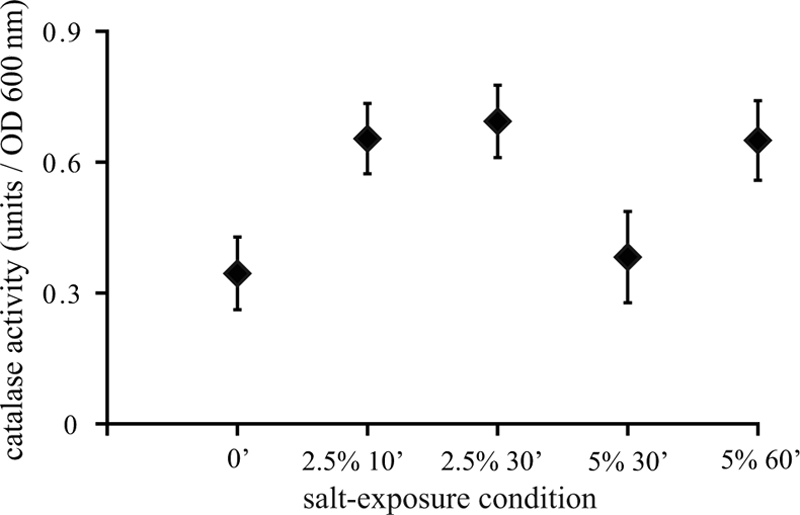
Catalase activities in exponentially growing cells and in cells exposed to 2.5% and 5% salt for 10 to 60 min. The error bars indicate standard errors.
Adaptation to 5% NaCl results in filamentation and reduced chemotaxis.
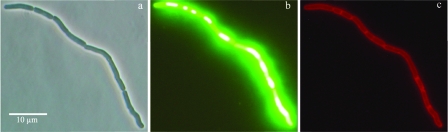
Exposure to 5% salt resulted in filamentation of cells, and the morphology of the filamentous cells was studied in more detail by using fluorescence microscopy. Cells exposed to 5% salt were stained using the green fluorescent nucleic acid stain SYTO-9 and FM 4-64, a red lipophilic dye for staining membranes. Figure 6a shows a phase-contrast image of filamentous cells exposed to 5% NaCl for 150 min, and Fig. 6b and c show cells labeled with SYTO-9 and FM 4-64. SYTO-9 stains the nucleoid inside the cell, and Fig. 6b shows that regularly spaced nucleoids were present in filamentous cells. Staining with the membrane stain FM 4-64 resulted in visible areas with dense red fluorescence, indicating that cell membrane septa were formed in the filamentous cells (Fig. 6c).
FIG. 6.
B. cereus ATCC 14579 exposed to 5% salt for 150 min: phase-contrast micrograph (a) and micrographs of cells labeled with SYTO-9 (a green fluorescent nucleic acid stain) (b) and FM 4-64 (a red fluorescent membrane stain) (c).
Several cell motility and chemotaxis genes were found to be downregulated upon exposure to 5% salt, and because cell motility and an active chemotactic machinery is required for chemotaxis (2), we tested the chemotactic behavior of cells exposed to 5% salt with the attractants trehalose and alanine. The chemotactic behavior of exponentially growing cells and that of cells exposed to 5% salt for 10 min and 60 min were comparable with both attractants (data not shown). However, prolonged exposure to 5% salt resulted in reduced chemotactic behavior with trehalose and alanine. The halo of the unstressed exponentially growing cells was denser than that of 5% salt-stressed cells, indicating the reduced chemotactic behavior of the latter type of cells (see Fig. S1 in the supplemental material).
NaCl stress induces cross-protection against heat and H2O2.
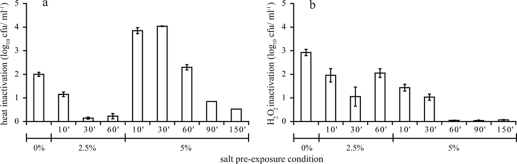
The transcriptome analyses of salt-stressed B. cereus ATCC 14579 cells revealed several features that could result in increased cellular resistance to other stress conditions. Therefore, the heat resistance and H2O2 resistance were determined for unstressed cells and for cells which were preexposed to 2.5% or 5% salt for 10 to 150 min. Cells preexposed to 2.5% salt (2.5% salt for 10 min, 2.5% salt for 30 min, and 2.5% salt for 60 min) were found to be more heat resistant than exponentially growing cells (time zero) (Fig. 7a). Cells exposed to 5% salt for 10 to 30 min were still in a lag phase, and these cells were very heat sensitive (5% salt for 10 min and 5% salt for 30 min). When 5% salt-stressed cells had resumed growth (5% NaCl for 60 min), the heat sensitivity decreased and was comparable to that of unstressed cells. Prolonged preexposure to 5% salt (5% salt for 90 min and 5% salt for 150 min) increased the heat resistance.
FIG. 7.
Inactivation of viable cells after heat treatment at 50°C for 2 min (a) or after exposure to 1 mM H2O2 for 1 min (b) for exponentially growing B. cereus ATCC 14579 cells and for cells which were preexposed to 2.5% and 5% for 10 to 150 min. Inactivation was determined by subtracting the log10 number of surviving cells after lethal treatment from the log10 number of cells at the start of the inactivation. The error bars indicate standard errors.
Preexposure to both mild and severe salt stress induced cross-protection against H2O2 stress (Fig. 7b). Cells exposed to 5% salt stress which had resumed growth (5% salt for 60 min, 5% salt for 90 min, and 5% salt for 150 min) were found to be the most H2O2-resistant cells.
DISCUSSION
Salt is a commonly used food additive, and bacteria can activate various cellular adaptation mechanisms to cope with salinity. In this study we described the phenotype and transcriptome responses of the pathogen B. cereus ATCC 14579 to salt stress. Two concentrations of added sodium chloride were used, 2.5% and 5%, in order to obtain insight into the overlap between the salt stress response and concentration-dependent parameters. Moreover, the use of various time frames for exposure to salt stress allowed adequate linkage of observed transcriptome responses to the phenotypic behavior of cells.
The transcriptome data confirmed that induction of osmoprotectant transporters is important in the mild and severe salt stress responses of B. cereus cells, which is in agreement with findings for other bacteria, such as B. subtilis, E. coli, and Listeria monocytogenes (21, 30). It has been demonstrated for B. subtilis that the substrate affinity of the glycine betaine ABC transporters OpuA and OpuC is in the same micromolar range as that of the glycine betaine symporter OpuD (20). However, the uptake via the ion-dependent symporters encoded by opuD and opuE seemed to be more favorable in nutrient-rich BHI medium than via ABC transporters, because the osmoprotectant ABC transporters were not differentially expressed in B. cereus ATCC 14579 upon salt stress. The gene encoding a putative di- and tripeptide transporter (BC0684) was also highly upregulated in B. cereus ATCC 14579 during exposure to both mild and severe salt stress. Previously, it has been demonstrated that a homologous gene (34%) in L. monocytogenes EGD-e (dtpT, LMO0555), encoding a di- and tripeptide transport system that supplies the cells with proline-containing peptides, is involved in salt stress protection (44). A similar function in the osmoregulation of B. cereus ATCC 14579 is conceivable as well. A transcriptome study of B. subtilis cells grown under high-salinity conditions showed that genes involved in the synthesis of the osmoprotectant proline were highly upregulated in this bacterium (33). However, induction of these genes was not observed in our study of the B. cereus ATCC 14579 salt stress response. This is consistent with previous investigations of de novo synthesis of compatible solutes in various Bacillus species (23), which demonstrated that proline was endogenously synthesized in osmotically stressed B. subtilis cells but not in B. cereus DSM31. In addition to proline, B. subtilis is also able to synthesize glycine betaine when the precursor choline is provided. A genome-wide search for orthologues of the gbsAB genes, which are known to be involved in glycine betaine synthesis in B. subtilis (4), revealed that these genes are not present in the genome of B. cereus ATCC 14579. These findings suggest that the osmoadaptation of B. cereus ATCC 14579 in BHI medium involves mainly the import of osmoprotectants rather than the synthesis of these compounds.
The upregulation of sigB and σB-dependent genes during exposure to both mild and severe salt stresses and the salt stress-induced translation of σB in B. cereus ATCC 14579 (35) suggested a role for σB during exposure to salt stress in this organism. For B. subtilis it has been shown that σB and σB-dependent proteins were strongly induced in response to salt stress at both the transcriptome (27) and proteome (15) levels. A sigB deletion mutant of B. subtilis was strongly impaired in osmotic stress adaptation (16, 39), as were a number of deletion mutants with mutations in σB-dependent genes (16). The osmoprotectant transporter OpuE was found to be σA and σB dependent in B. subtilis (32), indicating a physiological function in osmoregulation of a σB-dependent gene. In the current study, we demonstrated that the growth kinetics of exponentially growing cells of the B. cereus ATCC 14579 sigB deletion mutant FM1400 (35) and the rsbY deletion mutant FM1401 (36) after addition of salt were comparable to those of parental strain ATCC 14579. Previously, our laboratory showed that exposure to mild heat highly induced transcription of the σB regulon (37), and σB was found to be involved in the adaptive heat shock response of B. cereus ATCC 14579 (35). The set of σB-dependent genes in B. cereus ATCC 14579 appeared to be much smaller than that in other gram-positive species, such as B. subtilis and L. monocytogenes (37), and the σB regulon of this strain does not include genes encoding osmoprotectant transporters. Additionally, a large proportion of genes in the B. cereus ATCC 14579 σB regulon have unknown functions. Apparently, the role of σB and its regulon in the salt shock response of B. cereus ATCC 14579 is limited, although transcription and translation of σB are induced by salt stress.
Two putative CarD-like transcriptional regulators first described in Myxococcus xanthus (25) were highly expressed (BC3648 and BC4714) in B. cereus ATCC 14579 upon exposure to mild and severe salt stresses. To date, the information on CarD-like transcriptional regulators is limited. Recently, it was demonstrated that in Borrelia burgdorferi, a spirochete causing Lyme disease, a homologous gene for a CarD-like transcriptional regulator was expressed mainly during cultivation at a lower temperature (45). It is known that osmotic and chill stresses can trigger similar features in bacterial cells, indicating the overlap in the two responses (29, 41, 42). The role of the putative CarD-like transcriptional regulators in the stress response of B. cereus ATCC 14579 remains to be elucidated.
Exposure of exponentially growing cells to 5% salt resulted in an initial lag phase immediately after the salinity upshift, and filamentous cells were formed upon resumption of growth. Various bacteria have been shown to form filamentous cells under severe stress conditions (12, 19). The putative cell division inhibitor gene yfhF was found to be induced in B. subtilis upon exposure to salt stress (27), indicating that cell division might be affected. In this study, we showed that cell separation rather than septum formation was impaired in the filamentous B. cereus ATCC 14579 cells. Our transcriptome data revealed that genes encoding the FtsZ protein and the MinCD complex, which are known to be involved in ring formation at the midcell and correct septum placement (11), were not differentially expressed during exposure to severe salt stress in B. cereus ATCC 14579. On the other hand, genes involved in cell separation were downregulated, and these findings correspond to the inhibition of cell separation observed later when growth resumed.
Cell motility is required for chemotaxis toward attractants (2). The downregulation of genes involved in flagellum assembly and the chemotactic machinery during exposure to severe salt stress in this study suggested that the chemotactic behavior might be affected in the cells. Repression of genes and proteins involved in chemotaxis and motility was observed previously for B. subtilis exposed to high salinity (13, 15, 33) and also for B. cereus ATCC 14579 in response to bile salts (22). To test the chemotactic behavior of unstressed and salt-stressed cells, we used an assay in which we exposed high numbers of unstressed and salt-stressed cells to attractants in phosphate-buffered agar. This procedure allowed us to visualize the chemotactic behavior of B. cereus ATCC 14579 cells after brief incubation of the chemotactic plates, which resulted in increased cell density in the middle of the plate in the case of positive taxis. This chemotactic assay is growth rate independent because nutrients were not available in the agar and differences between unstressed and stressed cells could be observed in a short experimental time frame. We used this assay to demonstrate differences in the chemotactic behaviors of unstressed and salt-stressed cells because growth rate differences between unstressed and stressed cells may affect results obtained in cell motility assays which use nutritious media and longer incubation times to visualize chemotactic performance. The motility and chemotaxis genes were downregulated in cells exposed to 5% salt for 10 to 60 min, but the chemotactic behavior of these cells was not affected. This indicated that the cells were still able to sense and move toward attractants, despite the observed growth arrest in the severely salt-stressed cells. Therefore, it is conceivable that other cellular parameters contributed to the temporal growth inhibition. Prolonged exposure to 5% salt (150 min) resulted in reduced chemotaxis compared to that of unstressed cells, indicating that there was a temporal shift between the observed transcriptome responses of severely salt-stressed cells during the lag period and the phenotypic responses of these cells when growth resumed.
Our transcriptome data revealed that oxidative stress-related genes were upregulated during exposure to salt stress. Salt-induced expression of genes and proteins involved in the oxidative stress response was previously observed for B. subtilis (15, 27, 33). In our study, we demonstrated that upregulation of kat genes indeed resulted in increased catalase activity in salt-adapted cells compared to unstressed B. cereus ATCC 14579 cells. Cells adapting to severe salt stress, which were still in the lag phase, showed upregulation of kat genes, but the highest catalase activity and greatest resistance to H2O2 stress were observed in cells that had resumed growth during exposure to severe salt stress. Also, these findings demonstrate that there is a temporal shift between the transcriptome response of cells and development of the corresponding phenotype. In addition to salt-induced cross-protection against H2O2 stress, preexposure to salt induced cross-protection against heat. Aerobic heat stress has been demonstrated to impose an oxidative burden of reactive oxygen species in E. coli (3), and a recent study of E. coli showed that continuous osmotic and/or heat stresses induced upregulation of genes involved in the defense against oxidative stress damage (10). Therefore, it is conceivable that an activated oxidative stress response might confer cross-protection against other stresses. Furthermore, salt stress induced the transcription of osmoprotectant transporters, and it has been demonstrated that osmoprotectants (e.g., glycine betaine) can also provide protection against other stresses, such as heat (14) and chilling (41).
The whole-genome expression analyses of mildly and severely salt-stressed B. cereus cells revealed an overlap in the transcriptome responses. The so-called general salt stress transcriptome response involved activation of genes encoding osmoprotectant, Na+/H+, and di- and tripeptide transporters and an oxidative stress response. Activation of this general salt stress response may have contributed to the salt-induced cross-protection against heat and H2O2. Analysis of the transcriptome and phenotype responses in severely salt-stressed cells revealed a temporal shift between these responses for cellular filamentation, chemotactic performance, catalase activity, and optimal oxidative stress resistance. Therefore, comparison of mild and severe stress transcriptome profiles in combination with assessment of phenotypic characteristics can contribute to a better understanding of cellular stress adaptation strategies and possible cross-protection mechanisms.
Supplementary Material
Acknowledgments
We thank Natalie Commeau and Patrick P. L. A. de Leeuw for their assistance during the microarray experiments.
Footnotes
Published ahead of print on 24 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abee, T., and J. A. Wouters. 1999. Microbial stress response in minimal processing. Int. J. Food Microbiol. 50:65-91. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, S. I., I. B. Zhulin, L. Márquez-Magaña, and G. W. Ordal. 2002. Chemotaxis and motility, p. 437-452. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, DC.
- 3.Benov, L., and I. Fridovich. 1995. Superoxide dismutase protects against aerobic heat shock in Escherichia coli. J. Bacteriol. 177:3344-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boch, J., B. Kempf, R. Schmid, and E. Bremer. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178:5121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browne, N., and B. C. A. Dowds. 2001. Heat and salt stress in the food pathogen Bacillus cereus. J. Appl. Microbiol. 91:1085-1094. [DOI] [PubMed] [Google Scholar]
- 6.Carlin, F., H. Girardin, M. W. Peck, S. C. Stringer, G. C. Barker, A. Martinez, A. Fernandez, P. Fernandez, W. M. Waites, S. Movahedi, F. van Leusden, M. Nauta, R. Moezelaar, M. Del Torre, and S. Litman. 2000. Research on factors allowing a risk assessment of spore-forming pathogenic bacteria in cooked chilled foods containing vegetables: a FAIR collaborative project. Int. J. Food Microbiol. 60:117-135. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Besten, H. M. W., M. Mataragas, R. Moezelaar, T. Abee, and M. H. Zwietering. 2006. Quantification of the effects of salt stress and physiological state on thermotolerance of Bacillus cereus ATCC 10987 and ATCC 14579. Appl. Environ. Microbiol. 72:5884-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehling-Schulz, M., M. Fricker, and S. Scherer. 2004. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 48:479-487. [DOI] [PubMed] [Google Scholar]
- 10.Gunasekera, T. S., L. N. Csonka, and O. Paliy. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J. Bacteriol. 190:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precison. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 12.Hazeleger, W. C., M. Dalvoorde, and R. R. Beumer. 2006. Fluorescence microscopy of NaCl-stressed, elongated Salmonella and Listeria cells reveals the presence of septa in filaments. Int. J. Food Microbiol. 112:288-290. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann, T., A. Schütz, M. Brosius, A. Völker, U. Völker, and E. Bremer. 2002. High-salinity-induced iron limitation in Bacillus subtilis. J. Bacteriol. 184:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtmann, G., and E. Bremer. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höper, D., J. Bernhardt, and M. Hecker. 2006. Salt stress adaptation of Bacillus subtilis: a physiological proteomics approach. Proteomics 6:1550-1562. [DOI] [PubMed] [Google Scholar]
- 16.Höper, D., U. Völker, and M. Hecker. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsiao, A., T. Ideker, J. M. Olefsky, and S. Subramaniam. 2005. VAMPIRE microarray suite: a web-based platform for the interpretation of gene expression data. Nucleic Acids Res. 33:W627-W632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T., C. O. Gill, and L. M. McMullen. 2003. Behaviour of log-phase Escherichia coli at temperatures near the minimum for growth. Int. J. Food Microbiol. 88:55-61. [DOI] [PubMed] [Google Scholar]
- 20.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 22.Kristoffersen, S. M., S. Ravnum, N. J. Tourasse, O. A. Økstad, A. B. Kolstø, and W. Davies. 2007. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579. J. Bacteriol. 189:5302-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhlmann, A. U., and E. Bremer. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68:772-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morbach, S., and R. Krämer. 2002. Body shaping under water stress: osmosensing and osmoregulation of solute transport in bacteria. Chembiochem 3:384-397. [DOI] [PubMed] [Google Scholar]
- 25.Nicolás, F. J., R. M. Ruiz-Vázquez, and F. J. Murillo. 1994. A genetic link between light response and multicellular development in the bacterium Myxococcus xanthus. Genes Dev. 8:2375-2387. [DOI] [PubMed] [Google Scholar]
- 26.Periago, P. M., W. van Schaik, T. Abee, and J. A. Wouters. 2002. Identification of proteins involved in the heat stress response of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 68:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan, V. M., R. D. Sleator, G. F. Fitzgerald, and C. Hill. 2006. Heterologous expression of BetL, a betaine uptake system, enhances the stress tolerance of Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 72:2170-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sleator, R. D., and C. Hill. 2002. Bacterial osmoadaptation: role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 31.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 32.Spiegelhalter, F., and E. Bremer. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress-responsive promoters. Mol. Microbiol. 29:285-296. [DOI] [PubMed] [Google Scholar]
- 33.Steil, L., T. Hoffmann, I. Budde, U. Völker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenfors Arnesen, L. P., A. Fagerlund, and P. E. Granum. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32:579-606. [DOI] [PubMed] [Google Scholar]
- 35.Van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Schaik, W., M. H. Tempelaars, M. H. Zwietering, W. M. de Vos, and T. Abee. 2005. Analysis of the role of RsbV, RsbW, and RsbY in regulating σB activity in Bacillus cereus. J. Bacteriol. 187:5846-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Schaik, W., M. van der Voort, D. Molenaar, R. Moezelaar, W. M. de Vos, and T. Abee. 2007. Identification of the σB regulon of Bacillus cereus and conservation of σB-regulated genes in low-GC-content gram-positive bacteria. J. Bacteriol. 189:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Schaik, W., M. H. Zwietering, W. M. de Vos, and T. Abee. 2005. Deletion of the sigB gene in Bacillus cereus ATCC 14579 leads to hydrogen peroxide hyperresistance. Appl. Environ. Microbiol. 71:6427-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber, A., and K. Jung. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184:5502-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wemekamp-Kamphuis, H. H., R. D. Sleator, J. A. Wouters, C. Hill, and T. Abee. 2004. Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 70:2912-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wemekamp-Kamphuis, H. H., J. A. Wouters, R. D. Sleator, C. G. M. Gahan, C. Hill, and T. Abee. 2002. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl. Environ. Microbiol. 68:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wijnands, L. M., J. B. Dufrenne, F. M. Rombouts, P. H. in't Veld, and F. M. van Leusden. 2006. Prevalence of potentially pathogenic Bacillus cereus in food commodities in The Netherlands. J. Food Prot. 69:2587-2594. [DOI] [PubMed] [Google Scholar]
- 44.Wouters, J. A., T. Hain, A. Darji, E. Hüfner, H. Wemekamp-Kamphuis, T. Chakraborty, and T. Abee. 2005. Identification and characterization of di- and tripeptide transporter DtpT of Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 71:5771-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, X. F., M. S. Goldberg, M. He, H. Xu, J. S. Blevins, and M. V. Norgard. 2008. Differential expression of a putative CarD-like transcriptional regulator, LtpA, in Borrelia burgdorferi. Infect. Immun. 76:4439-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.