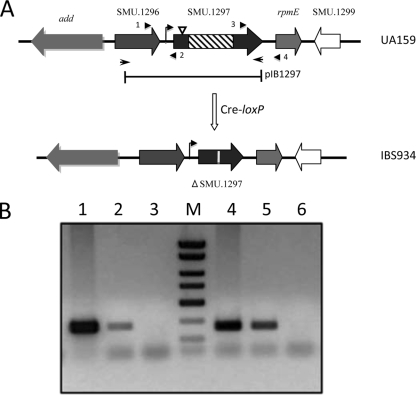
FIG. 2.
(A) Construction of a SMU.1297 deletion mutant. The direction of transcription of SMU.1297 and adjacent genes in the chromosome of S. mutans is indicated by block arrows. The SMU.1297 deletion strain, IBS934, was constructed using a Cre-loxP-based markerless gene deletion system (see Materials and Methods) that removed the central part of the SMU.1297 gene (hatched box), and inserted a 34-bp lox72 sequence to disrupt the gene (vertical white line). The bar below the chromosome indicates the region of SMU.1297 that was cloned into the complementing plasmid pIB1297. The site of the ISS1 insertion is shown by an inverted triangle. The bent arrow indicates a putative promoter region upstream of SMU.1297 gene. The diagram is not drawn to scale. (B) Transcriptional linkage analysis of the SMU.1297 locus. RNA, isolated from S. mutans UA159, was used as a template to produce cDNA, on which PCR was performed (lanes 2 and 5). RNA (lanes 3 and 6), and chromosomal DNA (lanes 1 and 4) were included as controls. Primers 1 and 2 (5′-GCCGGATCGACTGGACTGAAAGAATCGCTCAG-3′ and 5′-GCCCTCGAGCTGACTGCCTAATGCGTCAGGG-3′, respectively), shown by arrowheads in panel A, were used for the linkage analysis between SMU.1296 and SMU.1297, whereas primers 3 and 4 (5′-GGCCATCCATTAGCAAGTGGTG-3′ and 5′-TTGGTAACCCGTTGAAGTATCC-3′, respectively) were used for the linkage analysis between SMU.1297 and rpmE.