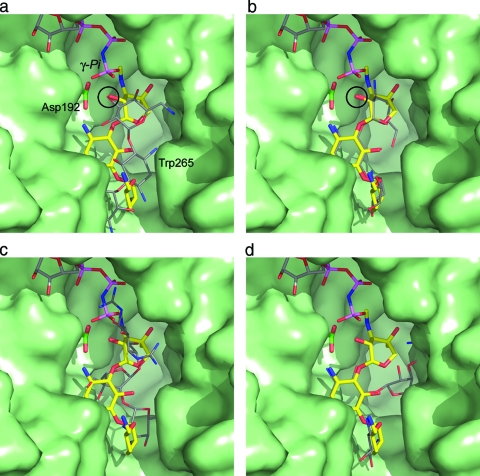
FIG. 5.
Surface representation of the substrate binding site in APH(2′′)-IIa (green) showing the bound gentamicin (yellow sticks) and an AMPPNP molecule (colored sticks; top) from APH(3′)-IIIa modeled into the structure based upon the superposition of the catalytic aspartate loop [residues 189 to 197 in APH(2′′)-IIa; 187 to 195 in APH(3′)-IIIa]. (a) The kanamycin from APH(3′)-IIa is shown as thin gray bonds. APH(3′)-IIa was superimposed on APH(2′′)-IIa based upon the catalytic aspartate loop. The location of this residue (Asp192) is indicated. When the structures are superimposed in this way, the hydroxyl group which is phosphorylated on both substrates is in essentially the same position (indicated by the black circle). (b) The kanamycin molecule modeled in the same conformation as gentamicin. In this orientation, the 2′′-OH of the kanamycin overlaps the equivalent atom on gentamicin (black circle). (c) The superimposed streptomycin from molecule B of the ternary AMPPCP-streptomycin-APH(2′′)-IIa complex, shown as thin gray sticks. The streptomycin occupies approximately the same location as the gentamicin. One of the guanidinium groups on streptomycin A ring projects upwards toward the nucleotide binding site and would clearly interfere with the β- and γ-phosphate groups of an extended ATP triphosphate. (d) A neomycin molecule modeled in the gentamicin binding site such that the A and B rings occupy approximately the same position as the corresponding rings in gentamicin. The C projects toward the helical subdomain, and in this orientation, the D ring of neomycin is hidden behind the helical subdomain.