Abstract
In the present study, we show in vitro binding of PrrA, a global regulator in Rhodobacter sphaeroides 2.4.1, to the PrrA site 2, within the RSP3361 locus. Specific binding, as shown by competition experiments, requires the phosphorylation of PrrA. The binding affinity of PrrA for site 2 was found to increase 4- to 10-fold when spermidine was added to the binding reaction. The presence of extracellular concentrations of spermidine in growing cultures of R. sphaeroides gave rise to a twofold increase in the expression of the photosynthesis genes pucB and pufB, as well as the RSP3361 gene, under aerobic growth conditions, as shown by the use of lacZ transcriptional fusions, and led to the production of light-harvesting spectral complexes. In addition, we show that negative supercoiling positively regulates the expression of the RSP3361 gene, as well as pucB. We show the importance of supercoiling through an evaluation of the regulation of gene expression in situ by supercoiling, in the case of the former gene, as well as using the DNA gyrase inhibitor novobiocin. We propose that polyamines and DNA supercoiling act synergistically to regulate expression of the RSP3361 gene, partly by affecting the affinity of PrrA binding to the PrrA site 2 within the RSP3361 gene.
Anoxygenic photosynthetic bacteria have been the subject of study for over 60 years (81). Investigations of the purple nonsulfur bacteria have contributed extensively to our understanding of redox control of gene expression as a mechanism used to maintain redox homeostasis (reviewed in references 3, 21, 45, and 56). Rhodobacter sphaeroides 2.4.1 is a purple nonsulfur bacterium which belongs to the Alpha-3-proteobacteria, whose members exhibit diversity in both their morphological and metabolic characteristics. With the availability of its genomic DNA sequence (http://www.rhodobacter.org) and the R. sphaeroides GeneChip (67), transcriptome and proteome analyses have been performed to investigate global cellular expression profiles in response to changes in growth conditions, as well as to the presence of specific mutations (1, 5, 6, 8, 9, 29, 61, 72, 89).
In order to maintain redox poise during adaptation to changing environmental conditions, R. sphaeroides controls gene expression through the interplay of discrete global regulatory systems. The principal players currently known in this genetic circuitry are the PrrBA two-component system (27, 28, 51), the Appa/PpsR antirepressor/repressor system (35, 69), and FnrL (88). PrrBA is a homolog of the RegBA two-component system of Rhodobacter capsulatus (23). PpsR is a repressor (61) homologous to CrtJ (22) of R. capsulatus, and FnrL (56) is homologous to Fnr (44) of Escherichia coli. The AppA antirepressor has no known homolog.
In the PrrBA system, PrrB is the histidine kinase/phosphatase and PrrA the response regulator. Previous data from this laboratory and others have shown regulation by PrrA, and RegA in R. capsulatus, of a considerable number of cellular functions; among these are photosynthesis, CO2 fixation, N2 fixation, H2 uptake and oxidation, and elements of the electron transport chain (21, 23, 56). These earlier observations were recently extended, upon finding that approximately 25% of the genome of R. sphaeroides is regulated by PrrA, either directly or indirectly, and that PrrA acts both as an activator and a repressor of transcription (29).
Similar to other two-component systems, with phosphorylation by its cognate histidine kinase/phosphatase PrrB (RegB) (27, 63, 77), PrrA (RegA) dimerizes (49) and binds to DNA using an H-T-H motif located at its carboxy-terminal end (19, 42, 48). As previously described, both unphosphorylated PrrA (U-PrrA) (12, 71) and unphosphorylated RegA* (U-RegA*) (19) are able to bind DNA in vitro and to activate transcription, although less efficiently than when they were phosphorylated (4, 20). It was also reported that U-RegA* binds to DNA targets with affinity similar to that with phosphorylated RegA* (P-RegA*), and these authors suggested that the phosphorylation state of RegA* might be relevant for transcription activation rather than DNA binding per se (38, 39). This might explain an in vivo effect of PrrA, since when prrA is present in multiple copies, expression of genes encoding proteins involved in photosynthesis occurs under aerobic growth conditions (28). Under these conditions, PrrA should not be phosphorylated, although other explanations are possible, such as PrrA being phosphorylated by other kinases, as shown previously for the HupT kinase when present in multiple copies (36).
In terms of the actual interaction with DNA, the consensus DNA binding sequences reported for PrrA (48, 58) and its homologs RegA, in R. capsulatus (76, 84), and RegR, in Bradyrhizobium japonicum (24) are highly degenerate, containing two DNA sites exhibiting imperfect dyad symmetry, with the quasisymmetrical sequences being separated by a variable spacer region containing anywhere from 0 to 10 nucleotides (nt) (58).
In addition to the exact identity, at the nucleotide level, of the DNA binding sequences for PrrA, the possible conformation of the DNA binding segment, as dictated by its sequence, has been suggested to influence binding of PrrA to DNA (48). In the case of the hemA1 promoter, it has been reported that the PrrA recognition sequences have the propensity to display a DNA conformation involving intrinsic high curvature (71), and indeed, PrrA itself displays some notable structural similarities to the DNA-bending FIS protein (48). There are also reports suggesting that DNA gyrase function, and hence the levels of DNA supercoiling, affects photosynthesis gene expression, which is positively regulated by PrrA (45), in both R. sphaeroides and R. capsulatus (53, 90).
In the present study, we investigated conditions for optimal in vitro binding by WT PrrA protein to a DNA binding sequence from R. sphaeroides. For this purpose, we chose PrrA site 2 (29) within the upstream regulatory region of the RSP3361 gene, a gene whose expression is positively regulated by PrrA and which contains two putative PrrA binding sites. Deletion of this particular binding sequence abolished PrrA regulation of the RSP3361 gene, and conversely, the absence of PrrA results in the absence of RSP3361 gene expression (29). Importantly, here, we show a requirement for PrrA phosphorylation to promote binding specificity to this DNA sequence, and we confirm this specificity by competition experiments. In an accompanying manuscript, we further investigate the binding specificity by conducting extensive mutagenesis of this DNA site (28a).
The presence of the polyamine spermidine in the binding reactions led to a marked increase in the binding affinity of phosphorylated PrrA to the RSP3361 locus PrrA site 2. Consistent with this, we found that the expression of the RSP3361 gene, as well as that of pucB (RSP0314 gene) and pufB (RSP6108 gene), two photosynthesis genes positively regulated by PrrA which encode the β polypeptides of the B800-B850 and B875 light-harvesting spectral complexes, respectively, was also positively regulated in the presence of spermidine, under aerobic growth conditions. In addition, the presence of spermidine in the growth medium increased the expression of additional photosynthesis genes, though modestly, as inferred by the production of light-harvesting spectral complexes under highly aerobic growth conditions. Thus, we provide evidence showing a correlation between the in vitro and in vivo spermidine effects in the case of the RSP3361 gene, and we show that these effects are presumably widespread to other PrrA-regulated genes.
In terms of the mechanism as to how spermidine affects the expression of the PrrA-regulated genes described here, two earlier lines of evidence describing (i) the strong effects of polyamines on DNA topology (31, 32, 46, 68, 80) and (ii) earlier suggestions about possible effects of DNA topology on PrrA-regulated gene expression (29, 48, 53, 71, 90) prompted us to test whether the observed effects of spermidine reported here might be due, at least in part, to changes in the topology of the DNA regulatory region promoted by the polyamine. We report that the expression of transcriptional lacZ fusions to the RSP3361 gene, as well as pucB (RSP0314 gene), is stimulated by negative supercoils, in vivo and in the absence of spermidine, as shown using transcriptionally driven topological alteration (TDTA) with a divergently transcribed wild-type (WT) (P) and mutant (Pdown) promoter to the RSP3361 gene regulatory region. This was confirmed using the DNA gyrase inhibitor novobiocin in the case of the RSP3361 gene.
Taken together, we provide evidence suggesting that expression of the RSP3361 gene, which is fully dependent on PrrA (29), is modulated by the topological state of the DNA and that polyamines, possibly due to their effects on DNA topology, act synergistically with DNA supercoiling to regulate expression of the RSP3361 gene, possibly by affecting phosphorylated PrrA (P-PrrA) binding.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains were grown at 37°C on LB medium (57) supplemented, when required, with the following antibiotics: tetracycline (Tet), at 20 μg/ml; streptomycin (St), at 50 μg/ml; spectinomycin (Sp), at 50 μg/ml; and ampicillin (Ap), at 150 μg/ml. R. sphaeroides 2.4.1 strains were grown at 30°C on Sistrom's medium A (SIS) (11) containing succinate as the carbon source, supplemented, as required, with the following antibiotics: Tet, at 1 μg/ml; St, at 50 μg/ml; Sp, at 50 μg/ml; and novobiocin (Nv), at 1 μg/ml. Anaerobic dark cultures were grown in SIS medium supplemented with 0.1% yeast extract in the presence of dimethyl sulfoxide (DMSO; 0.5% [vol/vol]), as an electron acceptor, and sparged with 95% N2-5% CO2. Aerobic cultures were grown in SIS medium and sparged with 30% O2, 69% N2, and 1% CO2. Spermidine trihydrochloride was used at 10 mM, compared to 3.8 mM NH4 and 34 mM succinate in SIS medium, for R. sphaeroides strains grown on plates, as well as in liquid cultures for the determination of light-harvesting spectral complexes and β-galactosidase activities. Plasmids were mobilized using triparental matings from E. coli DH5αPhe (28) into R. sphaeroides strains as described elsewhere (14).
DNA manipulations and analysis.
Standard protocols or manufacturer's instructions were used for the isolation of plasmid DNA, as well as for restriction endonuclease, DNA ligase, PCR, and other enzymatic treatments of plasmids and DNA fragments. Enzymes were purchased from New England Biolabs, Inc. (Beverly, MA), Promega Corp. (Madison, Wis.), United States Biochemical Corp. (Cleveland, OH), Invitrogen (Carlsbad, CA), and Roche (Branchburg, NJ). Plasmid DNA was purified using the Wizard SV miniprep kit from Promega (Madison, WI). Pfu ultra DNA polymerase was used as the high-fidelity PCR enzyme (Stratagene; Agilent Technologies, La Jolla, CA). DNA sequencing was performed at the DNA sequencing core facility of the Department of Microbiology and Molecular Genetics (The University of Texas—Health Science Center at Houston). The final versions of all relevant clones were sequenced to verify their construction.
PrrA purification and phosphorylation.
Typically, PrrA was purified from 1 liter of E. coli (JE4389) containing plasmid pPRRACHIS2, which contains the prrA gene with six-His codons immediately before the stop codon (63). A Ni-nitrilotriacetic acid His-Bind resin from Novagen (Madison, WI) was used for this purpose, following manufacturer's instructions. After elution in 20 mM Tris (pH 8.0), 0.5 M NaCl, and 0.5 M imidazole, the protein was dialyzed three times at 4°C over a period of approximately 18 h against buffer containing 20 mM Tris (pH 8.0), 200 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol (DTT), and 20% glycerol. The concentration of NaCl during the dialysis steps was successively decreased from 300 mM to 100 mM to no NaCl. After the final dialysis step, the concentration of glycerol was increased to 30%. The purified protein was concentrated using Centriplus YM-10 (10,000 molecular-weight cutoff) centrifugal filter units from Amicon (Millipore Corporation, Bedford, MA), by centrifuging at 4,500 rpm in a Sorvall SS-34 rotor at 4°C overnight, during which time the protein was typically concentrated from 4:1 to 5:1. The final glycerol concentration was increased to 40%, and the protein was resuspended in 50-μl aliquots and quick-frozen in a dry-ice/ethanol bath prior to storage at −80°C. The purification procedure yielded a protein that was ≥95% pure (data not shown). The bicinchoninic acid assay (Pierce, Rockford, IL) was used to determine protein concentration. Iodoacetamide was used to avoid interference due to the presence of the sulfhydryl reagent DTT in the sample.
PrrA phosphorylation was carried out by acetyl phosphate treatment, as described previously (12). A total of 25 μmol of purified PrrA, at a concentration of approximately 550 nmol/μl, were incubated at 30°C for 1 h, at a final concentration of 25 mM acetyl phosphate and 20 mM MgCl2. The binding reactions were performed immediately after completion of PrrA phosphorylation, and P-PrrA was prepared fresh for each experiment.
Labeling of DNA fragments.
pJE5096 is a p-Bend-3 derivative which contains the 16-nt RSP3361 gene PrrA site 2, and adjacent 40 nt on each side, as a PCR fragment cloned at its SalI site. Digestion of pJE5096 with EcoRV yielded a 191-bp fragment containing the RSP3361 PrrA site 2 at its center, with adjacent vector sequences. This EcoRV fragment was purified by electrophoresis twice, followed by gel extraction using the QIAquick gel extraction kit (Qiagen Inc., Santa Clarita, CA). The DNA fragments were then drop dialyzed against water for 1 h on 0.025-μm disc filters (Millipore, Billerica, MA) to eliminate impurities that could affect PrrA binding. DNA concentrations were calculated after reading the absorbance of several dilutions of the purified fragments at an optical density at 260 nm, using a UV-2450 UV-Vis spectrophotometer (Shimadzu, Kyoto, Japan).
DNA probes were made by end-labeling approximately 6 to 10 pmol of the appropriate 191-bp fragment containing the RSP3361 PrrA site 2 at an approximate concentration of 3 pmol/μl, with 5 μl of [γ-32P]ATP (Dupont NEN, Boston, MA). The final reaction volumes were 35 to 40 μl. T4 polynucleotide kinase (Invitrogen, Carlsbad, CA) was used for labeling, and the unincorporated nucleotides were removed using Micro Bio-Spin P-30 Tris chromatography columns (Bio-Rad, Hercules, CA), according to manufacturer's instructions, and the labeled DNA fragments were resuspended in water. After labeling and purification, aliquots of the [γ-32P]ATP-labeled DNA fragments were electrophoresed, and the dry gels were autoradiographed on BioMax XAR film (Kodak, Rochester, NY). The corresponding radioactive signals were quantitated as described previously (29).
PrrA binding and competition assays.
Binding and competition reactions were set up while the PrrA phosphorylation reactions were taking place, in order to have phosphorylated protein available when needed and to minimize the effects of possible instability at room temperature (RT) and spontaneous dephosphorylation. The mixtures were allowed to equilibrate to RT prior to initiating the reactions. The reactants (final concentrations) in a volume of 25 μl were, in order of addition, water, binding buffer (1×), 5 mM MgCl2, 0.05% Nonidet P-40 (NP-40), 2 mM spermidine trihydrochloride (when needed), ∼0.35 pmol labeled double-stranded 191-bp fragment containing the RSP3361 PrrA site 2, 80 ng/μl poly(dA) · poly(dT) nonspecific competitor DNA, and P-PrrA in storage buffer, at the appropriate concentrations. In some experiments, salmon sperm DNA was used at 67 ng/μl. The PrrA dilutions were done in 1× storage buffer. After the addition of P-PrrA, reaction mixtures were incubated at RT for 25 min, and immediately, 8 μl, approximately one-third, of each reaction mixture was loaded onto prerun gels, as described in the next section. For each experiment, both gels in the electrophoresis tank contained the same samples and, therefore, were run in duplicate. For competition experiments, the specific competitor DNA, at the appropriate concentration, was added after the P-PrrA addition. The 10× binding buffer contained 200 mM Tris (pH 7.9), 50 mM KCl, 5 mM MgCl2, and 1 mM DTT. The 1× PrrA storage buffer contained 40 mM Tris (pH 7.9), 50 mM KCl, 5 mM MgCl2, and 2 mM DTT, in 40% glycerol.
Electrophoretic mobility shift assays (EMSA).
Electrophoresis was performed in Mini-Protean 3 cells (Bio-Rad, Hercules, CA), using 7% gels made with a 40% stock of 29:1 acrylamide/bis-acrylamide and 0.5× Tris-borate-EDTA, as described previously (7), and with the following modification: both gels and running buffer had a final concentration of 200 μM spermidine in experiments where the polyamine was added to the binding/competition reactions. The apparent dissociation constant (Kapp) was calculated as described previously (16, 55) and corresponds to the concentration of P-PrrA that gave a fractional occupancy value of 0.5, or 50% occupancy of the DNA site. One hundred percent occupancy corresponds to a fractional occupancy of 1. The competition curve was generated by plotting unbound labeled DNA against increasing concentrations of the exact same unlabeled specific competitor DNA, using a constant amount of P-PrrA, in the appropriate reactions. The apparent inhibition constant (Ki) (33) was the concentration of unlabeled specific competitor DNA that displaced 50% of the labeled P-PrrA-bound DNA, corresponding to a fractional occupancy value of 0.5.
Transcriptionally driven topological alteration (TDTA).
Plasmids pJE5088 (this study) and pJE5090 (29) contain, respectively, an approximately 4.0-kb EcoRI insert from pML5PrrnB and pML5PdownrrnB, which are derivatives of the pML5 promoterless lacZ transcriptional fusion vector (47). This EcoRI DNA fragment harbors the multiple cloning site, most of the lacZ gene, and an upstream 336-bp fragment containing the divergently transcribed rrnB promoter (pJE5088) (18), or a mutated promoter (Pdown), made by an XmaI digest at a site within its spacer region, followed by Klenow fill-in. This modifies the 17-bp spacer region of the WT promoter to a promoter with 20 bp in the spacer region (Pdown). Promoters divergently transcribed from these two rrnB promoter alleles are expected to be subject to topological alterations in the form of negative supercoils, as predicted by the twin-domain model (54). The extent of negative supercoils incident on the RSP3361 gene promoter will vary proportionately as a function of the strength of the divergently transcribed promoter, in this case rrnB, as shown previously (65).
Determination of light-harvesting spectral complexes.
The amount of bacteriochlorophyll (Bchl) present in the B875 light-harvesting complex can be measured at 875 minus 820 nm (ɛ = 73 ± 2.5 mM−1 cm−1), normalized for 2 mol of Bchl per complex, whereas the concentration in the B800-B850 complex can be measured at 849 minus 900 nm (ɛ = 96 ± 4 mM−1 cm−1) (60).
β-Galactosidase assays.
Plasmid pCF200 (50) contains a transcriptional lacZ fusion to pucB (RSP0314 gene). pUI1663 and pUI1662 (25) harbor a transcriptional and a translational lacZ fusion to pufB (RSP6108 gene), respectively. pJE4935 and pJE4936 (29) contain transcriptional lacZ fusions to the RSP3361 gene (WT) and to a derivative of this gene with the PrrA site 1 deleted (29), respectively. Plasmids pJE5400 and pJE5403 contain, respectively, the WT rrnB and the rrnBPdown promoters divergently transcribed from pucB (RSP0314 gene), which was cloned as a PCR fragment containing 744 bp upstream pucB, up the PstI site, and 10 codons of the coding region. R. sphaeroides cultures used for the determination of β-galactosidase activity were grown as described previously (28), and assays were performed as described elsewhere (79). The data provided are the averages of the results of at least two separate experiments, each performed in duplicate. Standard deviations were always ≤15%. Protein concentration of cell extracts was determined using the bicinchoninic acid protein assay kit (Pierce, Rockford, IL), with bovine serum albumin as a standard.
Computer programs.
DNA analyses were performed using the computer programs ARTEMIS (Sanger Institute of The Wellcome Foundation) and DNA Strider (Institute de Recherche Fondamentale, Commissariat a l'Energie Atomique, France). Oligo 4.0 (National Biosciences Inc., Plymouth, MN) was used for primer designs. Bend.it, Plot.it, and Model.it, from DNAtools (ICGEB, Trieste, Italy), were used for the calculation of DNA bending and melting probabilities (83). WebSIDD (C. Benham, UC Davis Genome Center) was used for the calculation of the stress-induced (DNA) duplex destabilization (SIDD) profile (34) or the incremental free energy [G(x)] necessary to force a base pair at position x to be melted, represented as a plot of G(x) versus x. PREDICTOR (University of Sheffield, Department of Chemistry) was used for the calculation of the three-dimensional structure of double-stranded DNA (30). The EMBL gateway (http://www.embl-heidelberg.de/cgi/pi-wrapper.pl) was used for the calculation of isoelectric points. Microsoft Excel and Word (Microsoft), as well as Adobe Illustrator (Adobe), were used for manuscript preparation.
Materials.
5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) was purchased from USB Corporation, Cleveland, OH. o-Nitrophenyl-β-d-galactopyranoside (ONPG), lithium potassium acetyl phosphate (minimum 85%), spermidine trihydrochloride (minimum 98%), spermine tetrahydrochloride, salmon testes DNA and poly(dA) · poly(dT) were purchased from Sigma-Aldrich Corp., St. Louis, MO. GenePure LE agarose was purchased from ISC BioExpress, Kaysville, UT. Iodoacetamide was purchased from Pierce, Rockford, IL. NP-40 was purchased from Roche, Mannheim, Germany. All other chemicals used in this work were reagent grade.
RESULTS
In a previous study, we showed strong positive regulation by PrrA of the RSP3361 gene (29), whose expression appears to be entirely dependent on the presence of PrrA. Based on genomic annotation (http://www.rhodobacter.org), RSP3361 shows homology, at the amino-terminal end, to type I or type IV restriction endonucleases. The remainder of the protein shows homology to uncharacterized, conserved proteins and is accordingly assigned to COG4748, which contains uncharacterized, conserved proteins. Transcriptome analysis revealed that expression of this gene was minimal or absent under aerobic growth conditions and increased under anaerobic conditions, reaching maximal levels during photosynthetic growth at 10 W/m2 incident light intensity. The induction of the RSP3361 gene was confirmed by direct mRNA analysis, as well as reporter fusion experiments (29). Whatever function the RSP3361 protein performs, there is an apparently greater cellular requirement when R. sphaeroides grows under anaerobic light conditions, and this expression tends to mimic photosynthesis gene expression although the expression of this gene is not dependent upon any of the other known regulators of photosynthesis gene expression.
Regulatory and structural elements in the regulatory region of the RSP3361 gene.
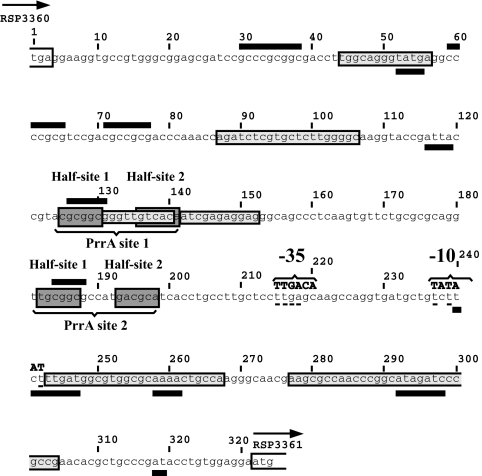
The regulatory region of the RSP3361 gene is depicted in Fig. 1. Two putative PrrA binding sequences (58) were found upstream of a putative σ70-type promoter (59). These putative PrrA sites encompass two half-sites with strong dyad symmetry, separated by a spacer region containing 5 nt. We have shown previously, by reporter fusion analysis, that deletion of the 16 nt encompassing either PrrA sites 1 or 2 decreases expression of the RSP3361 gene by approximately 37% or completely abolishes the induction of this gene, respectively (29).
FIG. 1.
Regulatory region of the RSP3361 gene. The numbered nucleotides in the intergenic region between the RSP3360 gene and the RSP3361 gene are shown. Both presumptive PrrA sites are indicated, with their respective half-sites (1 and 2) marked by dark-shaded boxes. The size of half-site 2 is increased from 5 to 6 nt, and the data justifying that change are shown in the accompanying paper (28a). The putative σ70 promoter binding sequence is indicated in boldface type, above the sequence, and the matching nucleotides in the −35 and −10 regions are underlined. DNA regions with intrinsic bending propensity are highlighted by light-shaded boxes. Areas requiring higher energy to melt, according to SIDD analysis, are denoted by thick lines above the sequences, whereas areas requiring low energy are shown by thick lines below the sequences. The direction of transcription for both genes is indicated by arrows. The RSP3360 gene, encoding a putative adenine-specific methyltransferase, is transcribed in the same direction as is the RSP3361 gene, but is not PrrA regulated (29).
In addition to the PrrA sites indicated, a good match was found for a presumptive σ70-type promoter; this site contains −35 and −10 regions and 16-nt spacer regions, similar to the canonical σ70 binding site (59), and is depicted in Fig. 1, although σ70 binding has not been demonstrated. The absence of other known consensus DNA binding sequences in this regulatory region is consistent with, although it does not prove, the apparent, strict dependency of RSP3361 gene expression on PrrA (29). This apparent, strict dependency on PrrA makes this locus ideal for the studies described herein, unlike many of the photosynthesis genes, which have multiple regulator interactions (6, 56).
We used the bioinformatics tools described in Materials and Methods to detect DNA sequences upstream of the RSP3361 gene with a tendency for intrinsic bending (83) or to become single stranded through SIDD (34). Five DNA regions were found, ranging in size from 13 to 25 nt, with intrinsic propensity for a high degree of curvature. The potential for PrrA binding sites to possess intrinsic curvature has been suggested in earlier studies (71), but none of these sequences overlapped the PrrA site studied here.
DNA sequences with the lowest probability to become single stranded due to supercoiling energy (34) were found upstream of the apparent promoter region; two such regions overlapped half-site 1 in both PrrA binding sequences. Alternatively, sequences with the highest melting probability clustered downstream from the promoter, and one of these, an 8-nt region, overlapped the putative −10 region and immediate downstream sequences. The presence of a DNA sequence with a high tendency to become single stranded, overlapping the −10 region of the putative promoter, reinforces our choice of this promoter, which was based on its sequence and position with respect to the coding region of the gene. This structure also suggests the possibility for this −10 region to progress from closed to open complex form by use of supercoiling energy.
Effects of phosphorylation and nonspecific competitor DNA concentration on PrrA binding to the RSP3361 gene PrrA site 2.
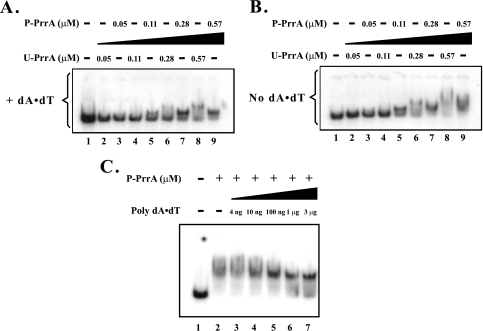
We purified and phosphorylated PrrA, as described in Materials and Methods and in a previous study (12), and tested the effects on DNA binding by adding increasing amounts of U-PrrA and P-PrrA to the 191-bp DNA fragment containing the RSP3361 gene PrrA site 2, which when deleted, leads to the absence of RSP3361 gene expression (29). The results of the EMSA are shown in Fig. 2. P-PrrA, but not U-PrrA, formed a specific, stable DNA-protein complex (Fig. 2A). U-PrrA, although found to bind to this DNA sequence, did so in a nonspecific manner, as inferred from the large size of the U-PrrA-DNA complex, compared to that of the P-PrrA-DNA complex, at the same U-PrrA and P-PrrA concentrations (Fig. 2A, lanes 8 and 9). As expected, in the absence of the nonspecific competitive inhibitor poly(dA) · poly(dT), which was used at a final concentration of 80 ng/μl (2 μg total), more nonspecific binding was shown for P-PrrA, as evidenced by the very diffuse gel signals (Fig. 2B), leading to less-stable DNA-protein complexes. Thus, PrrA phosphorylation is apparently required for specific binding to the DNA fragment containing the RSP3361 gene PrrA site 2 in vitro.
FIG. 2.
Effects of PrrA phosphorylation and nonspecific competitor DNA addition on PrrA binding properties. EMSA results showing binding by increasing concentrations of either P-PrrA (lanes 3, 5, 7, and 9) or U-PrrA (lanes 2, 4, 6, and 8) to RSP3361 gene PrrA site 2. The analysis is performed in the presence (A) and absence (B) of the nonspecific competitive inhibitor poly(dA) · poly(dT), which was added at an 80 ng/μl concentration. (C) Effects of increasing concentrations of poly(dA) · poly(dT) on P-PrrA-DNA complex formation at a constant P-PrrA concentration. The amounts shown in each case refer to the total amount added to the reaction. The plus sign refers to addition of 0.57 μmol of P-PrrA. In all panels, lane 1 contains DNA only. The minus sign indicates that there was no addition of the specific compound.
The above poly(dA) · poly(dT) concentration (80 ng/μl) was chosen since approximately the same amount of P-PrrA-DNA complex was formed using either 1 μg (40 ng/μl) (Fig. 2C, lane 6) or 3 μg (120 ng/μl) (Fig. 2C, lane 7) of poly(dA) · poly(dT).
Effect of spermidine on PrrA binding to the RSP3361 gene.
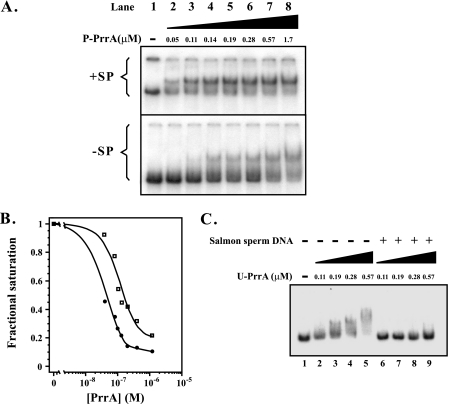
Once we optimized for the nonspecific competitor DNA concentration, we investigated the effects of additional factors found to increase the binding affinity of other transcriptional regulators. One of these was polyamines, which have been reported to increase the binding specificity of other transcriptional factors (66). To determine whether the affinity of purified PrrA for PrrA site 2, upstream of the RSP3361 gene, was affected by the presence of the polyamine in vitro, we tested binding by the addition of increasing amounts of P-PrrA to the RSP3361 gene PrrA site 2 in the presence and absence of spermidine. The results are shown in Fig. 3. In the absence of spermidine, there was no appreciable binding of P-PrrA to DNA at the lowest protein concentration (∼55 nM) used in the experiment. Spermidine addition resulted in enhanced binding, as evidenced by P-PrrA-DNA complex formation (Fig. 3A). The amount of spermidine used in these experiments was determined experimentally using different concentrations, and similar results were obtained using the polyamine spermine (data not shown).
FIG. 3.
Effect of spermidine on P-PrrA and U-PrrA binding affinity in vitro. (A) EMSA results showing binding of increasing concentrations of P-PrrA to an ∼0.35-pmol labeled double-stranded 191-bp fragment containing the RSP3361 PrrA site 2. The reaction conditions are described in Materials and Methods. The analysis was performed in the presence (+SP) and absence (−SP) of 2 mM spermidine. Both gels were run identically. PrrA concentrations (μM) are indicated above the lanes. (B) Quantitation of unbound DNA as a function of P-PrrA concentration. The calculated values of 4.6 × 10−8 M (+SP) and 1.9 × 10−7 M (−SP) correspond to the P-PrrA concentrations at which 50% of the labeled DNA is unbound, or 0.5 fractional saturation. Poly(dA) · poly(dT) was added as a nonspecific competitor at 80 ng/μl concentration. (C) Effects of increasing concentrations of U-PrrA on U-PrrA-DNA complex formation in the presence or absence of salmon sperm DNA, with 2 mM spermidine added to the reactions. The plus sign refers to addition of 2 μg of salmon sperm DNA (67 ng/μl). The concentrations of U-PrrA (μM) are indicated above the lanes. In all panels, lane 1 contains DNA only. The minus sign indicates that there was no addition of the specific compound.
P-PrrA-DNA complexes formed in the absence of spermidine were diffuse and difficult to quantitate, possibly due to complex instability. Thus, the data presented in Fig. 3B correspond to the estimation of unbound DNA, rather than bound DNA, as a function of P-PrrA concentration, which are not true dissociation constants (see next section). The values corresponding to the concentrations of P-PrrA at which 50% of the labeled DNA is in a noncomplexed form, or to a fractional saturation of 0.5, were ∼4.6 × 10−8 M and ∼1.9 × 10−7 M, in the presence and absence of spermidine, respectively. Visual inspection of these and additional autoradiographed gels (data not shown) indicated that ∼50% of the DNA was consistently complexed with ∼55 nmol of P-PrrA (Fig. 3A, lane 2), in the presence of spermidine, and with ∼570 nmol of P-PrrA (Fig. 3A, lane 7), in its absence—an ∼10-fold difference in P-PrrA concentration. Thus, the calculated approximately fourfold difference in concentrations of P-PrrA at which 50% of the labeled DNA is in a noncomplexed form with PrrA in the presence and absence of spermidine is most likely an underestimation as the result of the diffuse nature of the complex in the absence of spermidine, and the difference could be as high as 10-fold.
In the absence of the polyamine (Fig. 3A, bottom panel), the rate of gel migration of all DNA species, free and bound, was higher, as evidenced by the differences in the distances separating these species from the loading wells. This probably reflects the fact that polyamines, which were present in the gel and the running buffer in the experiment depicted in the top panel, affect the structure of the DNA fragment and, therefore, improve the binding of P-PrrA to its DNA site (see Discussion). However, it cannot be ruled out that these polycations have a neutralizing electrostatic effect on the overall charge of the DNA fragments and, therefore, slow down their migration toward the anode.
Binding of U-PrrA to RSP3361 PrrA site 2 in the presence of spermidine (Fig. 3C, lanes 2 to 5) was prevented by the addition of 2 μg (67 ng/μl final concentration) of nonspecific competitor salmon sperm DNA, as depicted in Fig. 3C, lanes 6 to 9.
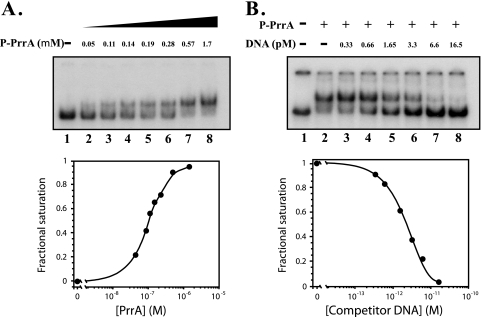
To demonstrate that phosphorylation of PrrA is required for achieving specific binding to the RSP3361 gene PrrA site 2, as well as to show that the binding of P-PrrA was specific, we conducted DNA binding experiments, using increasing amounts of P-PrrA, to a DNA fragment containing the RSP3361 gene PrrA site 2 (Fig. 4A), and we tested the effect of adding increasing amounts of unlabeled specific competitor DNA to a preformed P-PrrA-labeled DNA complex in the presence of spermidine (Fig. 4B). By quantitating the intensities of P-PrrA-DNA complexes, we calculated an apparent dissociation constant, or Kapp, equal to ∼1.3 × 10−7 M (Fig. 4A, bottom panel). The results of the competition experiment are shown in Fig. 4B. A constant amount of P-PrrA, ∼0.57 μmol, complexed with the radioactive RSP3361 gene fragment was displaced by increasing amounts of a double-stranded, unlabeled 191-bp fragment containing the RSP3361 PrrA site 2. We calculated the inhibitory constant, or Ki (33), to be approximately 2.54 × 10−12 M. This value indicates the concentration of competitor DNA required to effect the displacement of P-PrrA from 50% of the labeled DNA in a P-PrrA-DNA complex and corresponds to an ∼7.8-fold molar excess of the competitor unlabeled DNA, with respect to the labeled DNA.
FIG. 4.
Determination of the affinity of P-PrrA for PrrA site 2 in the RSP3361 gene. (Top panels) Autoradiograms of the EMSA experiments. (Bottom panels) Quantitation. (A) Lane 1 contains a labeled 191-bp fragment containing the RSP3361 PrrA site 2 only. A calculated Kapp of 1.3 × 10−7 M corresponds to 50% occupancy, or 0.5 fractional saturation. (B) Approximately 0.35 pmol of the labeled double-stranded 191-bp fragment containing the RSP3361 PrrA site 2 was incubated with ∼0.57 μmol of P-PrrA, equivalent to the amount of P-PrrA used in lane 7 in panel A (top). In addition, increasing amounts of the double-stranded unlabeled 191-bp fragment containing the RSP3361 PrrA site 2 were added as specific competitor DNA. The competitor DNA amounts added are shown, and they constitute 1× (lane 3), 2× (lane 4), 5× (lane 5), 10× (lane 6), 20× (lane 7), and 50× (lane 8), relative to labeled DNA. Lane 1 contains a labeled 191-bp fragment containing the RSP3361 PrrA site 2 only, and lane 2 contains the labeled fragment and P-PrrA. A Ki value of 2.54 × 10−12 M indicates 50% displacement of P-PrrA, and it corresponds to ∼7.8×, relative to 1× the labeled DNA fragment.
Generalized and localized supercoiling effects on expression of the RSP3361 gene.
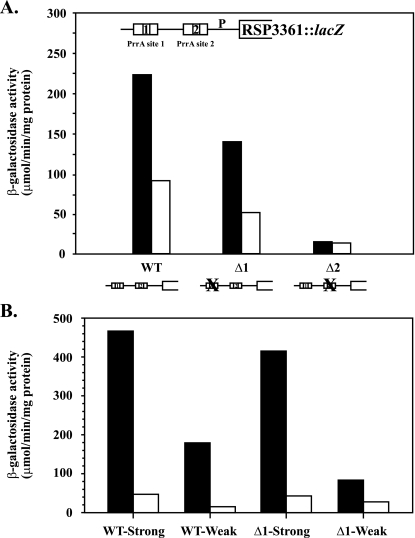
Because of previously reported effects of polyamines on DNA topology (31, 32, 46, 68, 80) and earlier suggestions about the possible effects of DNA topology on PrrA-regulated gene expression (29, 48, 53, 71, 90), we chose to test the possible effects of DNA supercoiling on expression of the RSP3361 gene. DNA supercoiling has well-known generalized effects on gene expression (70) and on photosynthesis genes, which are mostly PrrA regulated, in particular (53). To this effect, we decided to test the β-galactosidase activity of Φ(RSP3361-lacZ) (29) in WT R. sphaeroides strains grown in the presence and absence of the DNA gyrase inhibitor Nv. The expression of the transcriptional fusion decreased by approximately 50% in the presence of the inhibitor, as shown in Fig. 5A. The same was true for a fusion containing a deletion of the entire PrrA site 1, whereas deletion of PrrA site 2 completely eliminated expression, as had been previously shown (29). Thus, expression of the RSP3361 gene is partly dependent on DNA supercoiling levels and favored by negative supercoiling.
FIG. 5.
Supercoiling effects on expression of the RSP3361 gene. (A) β-Galactosidase activities of Φ(RSP3361-lacZ) (pJE4935 [29]) contained in vector pML5 (47) in WT R. sphaeroides grown under anaerobic growth conditions in the dark with DMSO. Black bars, without Nv; white bars, with Nv. WT at the bottom represents the WT fusion, whereas Δ1 and Δ2 represent, respectively, fusions deleted for the upstream (PrrA site 1) and downstream (PrrA site 2) PrrA sites, which have been described previously (29). (B) β-Galactosidase activities of Φ(RSP3361-lacZ) contained in a version of pML5 derived from pJE5088 and pJE5090. pJE5088 contains the rrnB WT promoter (designated as WT-Strong) divergently transcribed from the RSP3361 gene promoter, whereas pJE5090 contains the mutated version of rrnB Pdown (designated as WT-Weak), divergently transcribed from the RSP3361 gene promoter. WT (black bars) and PrrA2 (white bars) R. sphaeroides cells containing these plasmids were grown under anaerobic growth conditions in the dark and with DMSO. WT at the bottom represents the WT Φ(RSP3361-lacZ), whereas Δ1 represents a Φ(RSP3361-lacZ) deleted for the upstream PrrA site, PrrA site 1. Thus, WT-Strong and Δ1-Strong contain the divergently transcribed WT rrnB promoter from either WT Φ(RSP3361-lacZ) or Φ(RSP3361Δ1-lacZ), respectively, and WT-Weak and Δ1-Weak containing the rrnB Pdown also divergently transcribed for both respective Φ(RSP3361-lacZ).
Since DNA gyrase inhibitors show notoriously pleiotropic effects on cells, we constructed lacZ transcriptional fusion vectors harboring either the strong rrnB R. sphaeroides promoter (18) or a mutated version, as explained in Materials and Methods, divergently transcribed from the target RSP3361 gene promoter. Thus, negative supercoils arising from transcription by RNA polymerase at these two promoter alleles, as predicted by the twin-domain model (54), should vary as a function of rrnB promoter strength, as shown previously for other promoters (65), and affect RSP3361 gene expression accordingly. Since the rrnB promoters are divergently transcribed from the gene of interest, runaway transcription from these promoters affecting the experimental results is not a concern.
Expression of the Φ(RSP3361-lacZ) transcriptional fusion was approximately 2.5-fold greater when the WT rrnB promoter (WT-Strong) was used instead of the Pdown allele (WT-Weak) in cells grown anaerobically in the dark with DMSO as the electron acceptor, as shown in Fig. 5B. This was consistent with the result obtained using the DNA gyrase inhibitor and confirms that this promoter is positively regulated by negative supercoiling. The same was true for the RSP3361 gene promoter containing the deletion of PrrA site 1 (29), although in this case the difference was greater, by about fivefold. In addition, RSP3361 gene expression was fully dependent on PrrA, as reported previously (29).
PrrA regulation of polyamine biosynthesis and transport.
Because of the effects of spermidine, as observed here, we examined our previous microarray data comparing global gene expression in a PrrA mutant (PrrA2) to the WT (29), and we observed the apparent regulation by PrrA of genes involved in polyamine metabolism. Biogenic amines have been previously shown to have pleiotropic effects on bacterial DNA metabolism (85). Specifically, speC (RSP1991 gene), which encodes l-ornithine decarboxylase (SpeC) (EC 4.1.1.17), was found to be positively regulated by PrrA, by approximately 4.2-fold. This enzyme is induced under anaerobic growth conditions in other prokaryotes (reviewed in reference 78). The genes encoding l-arginine decarboxylase (SpeA) and S-adenosylmethionine decarboxylase (SpeD), the rate-limiting enzymes in alternative polyamine biosynthetic pathways in other organisms (reviewed in reference 78), are apparently absent in R. sphaeroides, as inferred from gene annotation (http://www.rhodobacter.org). Similar to polyamine biosynthesis, the transport of polyamines was also found to be regulated by PrrA; as many as six different ABC transporter systems with specificity for polyamines, as inferred from gene annotation, were, for the most part, positively regulated by PrrA (29). Based upon these inferred regulatory effects on biogenic amine metabolism, we tested for the effect of polyamines on PrrA binding, as described earlier, but in addition, measured the effects of polyamines in situ on growing cells of R. sphaeroides.
Induction of the photosynthetic apparatus under aerobic growth conditions by spermidine.
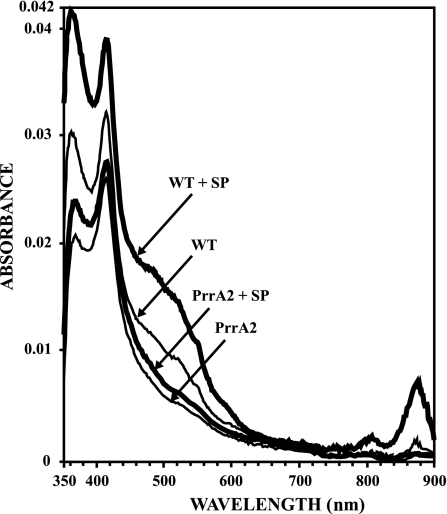
To assess the effect of spermidine in vivo we grew R. sphaeroides WT cells under highly oxygenic conditions, as described in Materials and Methods, and measured the Bchl levels associated with the light-harvesting spectral complexes (60). The results are shown in Fig. 6. In the presence of 10 mM spermidine in the growth medium, WT cells showed an approximately fivefold increase in Bchl associated with the light-harvesting complex I (LHI), or B875, relative to that shown by cells growing in media with no spermidine supplementation. These results are similar to our earlier results showing induction of the photosynthetic apparatus under aerobic growth conditions, either in the presence of multiple copies (∼5 to 7) of PrrA (27) or upon deletion of the oxidoreductase PrrC (26) or the cbb3 subunit CcoQ (62). The PrrA mutant, PrrA2 (27), was used as a negative control in this experiment, since PrrA positively regulates most genes encoding proteins involved in photosynthesis (45). As expected, PrrA2 mutant cells did not produce light-harvesting spectral complexes in the presence or absence of spermidine. This result can be interpreted as demonstrating a cause and effect between polyamines and photosynthesis gene expression under aerobic conditions.
FIG. 6.
Light-harvesting spectral complex formation in the presence of spermidine. Cells were grown sparged with a mixture of 30% O2, 69% N2, and 1% CO2 in Sistrom's medium and harvested at a low cell density of approximately 0.14 absorbance units at an optical density at 600 nm to prevent oxygen limitation due to cellular growth. The protein concentration is the same for all samples represented. SP denotes spermidine, which was used at 10 mM concentration in the growth media. PrrA2 is a PrrA mutant used in our laboratory (27).
In order to test whether the production of light-harvesting spectral complexes was, in part, a direct result of the effect of polyamines on gene transcription, we used reporter fusions to genes encoding polypeptides of the photosynthetic apparatus. The results are shown in Table 1. Consistent with our previous results, we found an approximately twofold increase in the expression of pucB (RSP0314 gene), pufB (RSP6108 gene), and the RSP3361 gene. In the case of the last gene, this effect was observed either for the WT fusion, containing two PrrA sites, or for a fusion in which PrrA site 1 was deleted. The presence of spermidine in the growth media also led to an approximately twofold increase in expression of a translational fusion to pufB. This increase in gene expression while growing in the presence of spermidine occurred under aerobic growth conditions, but not anaerobically in the dark in the presence of DMSO, where expression was indistinguishable in the presence and absence of the polyamine (data not shown).
TABLE 1.
Effect of spermidine on the β-galactosidase activity of lacZ fusions to pucB, pufB, and the RSP3361 genec
| lacZ fusiona | Vectorb | Spermidine
|
|
|---|---|---|---|
| Absent | Present | ||
| Φ(pucB-lacZ) | pCF200 | 272.4 | 517.5 |
| Φ(pufB-lacZ) | pUI1663 | 398.3 | 506.5 |
| Φ(pufB-lacZ) | pUI1662 | 36.4 | 62.0 |
| Φ(RSP3361-lacZ) | pJE4935 | 9.8 | 24.6 |
| Φ(RSP3361Δ1-lacZ) | pJE4936 | 2.6 | 6.8 |
| rrnB(div.)-Φ(pucB-lacZ) | pJE5400 | 71.3 | 177.7 |
| rrnBPdown(div.)-Φ(pucB-lacZ) | pJE5403 | 24.5 | 51.9 |
“(div.)” indicates that the rrnB promoters are divergently transcribed from pucB. Δ1 indicates a deletion of PrrA site 1.
The lacZ fusion vectors used were constructed as described in the following references: pCF200 (50); pUI1663 and pUI1662 (25); pJE4935 and pJE4936 (29); pJE5400 and pJE5403 (this study). All fusions are transcriptional, except for that of pUI1662, which is a translational fusion to pufB.
Cells were grown aerobically by sparging with 69% N2, 30% O2, and 1% CO2. Units of β-galactosidase are expressed in μmol/min/mg protein. Experiments were performed in duplicate, and the standard deviation was ≤15%.
DISCUSSION
We have analyzed expression of the RSP3361 gene of R. sphaeroides (this study; 28a, 29), which appears to be entirely dependent upon P-PrrA, unlike many of the photosynthesis genes, which show more complex regulatory interactions (6, 56). The data presented here show specific binding of P-PrrA in vitro to PrrA site 2 of the RSP3361 gene regulatory region. Similar to what has been reported in other studies and alluded to earlier, U-PrrA was found to bind to the DNA fragment containing the RSP3361 gene PrrA site 2, but the binding was shown to be nonspecific, since no U-PrrA-DNA complex was observed upon including an appropriate concentration of nonspecific competitor DNA, and U-PrrA-DNA complexes increased in size concomitantly to increases in protein concentration, as observed by EMSAs. Whether the observed U-PrrA nonspecific binding occurs at RSP3361 gene PrrA site 2 and/or elsewhere within the DNA fragment used in these experiments is not known.
Although we have demonstrated a requirement for PrrA phosphorylation to impart binding specificity toward RSP3361 gene PrrA site 2, the possibility still remains for U-PrrA to specifically bind to different DNA sites in the genome, as suggested before for both PrrA and RegA, in R. sphaeroides and R. capsulatus, respectively (4, 12, 19, 20, 38, 39, 71), and by our own earlier data describing photosynthesis gene expression aerobically, when prrA (RSP1518 gene) was in multiple copies in R. sphaeroides (28).
The P-PrrA binding specificity to the RSP3361 gene PrrA site 2 was demonstrated by the addition of nonspecific competitor DNA, which does not affect P-PrrA binding, in contrast to U-PrrA, and by competition kinetics with specific competitor DNA. In a separate study (28a), we found similar binding kinetics for three other P-PrrA binding sites in the genome. The DNA sequence of this P-PrrA-binding site is T-G-C-G-G-C-N(5)-G-A-C-G-C-A, encompassing two half-sites containing 6 nt, separated by a 5-nt spacer. Near-perfect symmetry is displayed between both half-sites, except for position 5. As stated previously (29), this site shows a perfect match to a published consensus for PrrA binding sequences (58), but we have extended the length of half-site 2 from 5 nt, as reported in that study (58), to 6 nt, since mutation of this added nucleotide abolishes P-PrrA binding to the PrrA site 2 in the RSP3361 gene (28a).
While setting up the in vitro binding assays for PrrA and its DNA targets, we found that the affinity of P-PrrA binding for RSP3361 gene PrrA site 2 in vitro increased 4- to 10-fold when the polyamines, spermidine and spermine, were present in the binding reaction, similar to what has been reported for other DNA binding proteins (see below). Since both polyamines affected binding of P-PrrA to this DNA site, we hypothesize that the polyamine effects are likely due to their multivalent cationic nature. Polyamines are present intracellularly at millimolar concentrations in both prokaryotes (∼6 mM spermidine in E. coli [78] and photosynthetic bacteria [37]), and eukaryotes (10) and have also been reported to affect transcription (43) and translation (87) in prokaryotes as well as in eukaryotes (2, 66, 73).
Transcriptional lacZ fusions to the photosynthesis genes pucB (RSP0314 gene) and pufB (RSP6108 gene) and the RSP3361 gene, as well as a translational lacZ fusion to pufB, revealed a twofold increase in expression due to the presence of spermidine when R. sphaeroides cells were grown aerobically, and this was reflected by the modest production of light-harvesting spectral complexes, albeit comparable to that found in previous studies (26, 62). This suggests that the production of light-harvesting spectral complexes under aerobic growth conditions might be due to a combination of transcriptional and posttranscriptional effects (87). Likely, the extracellular spermidine concentration used in our experiments might have resulted in an at least transiently higher intracellular polyamine concentration, as previously reported for other organisms (74, 82; also as reviewed in references 2, 41, and 78). The fact that the spermidine effects were only observed when the cells grew aerobically might be a reflection of either differential polyamine biosynthesis and transport depending on growth conditions, as inferred from microarray expression data (29) and/or the fact that slight effects on gene regulation and light-harvesting spectral complex formation might not be observed against the background of high photosynthesis gene expression under anaerobic conditions.
The simplest mechanistic interpretation for how polyamines might be affecting P-PrrA binding to DNA as well as PrrA-regulated gene expression would be that, by their cationic nature, they might simply be neutralizing the negative charge of the DNA backbone, as opposed to favoring specific electrostatic interactions. In this respect, the amino acids in the HTH motif of PrrA predicted to make specific contacts with the DNA major groove (42, 48) are R171, R172, Q175, and R176, whereas R143, R158, N159, and R171 have been proposed to contact the DNA phosphate backbone and possibly be involved in nonspecific interactions (48). Alternatively, or in conjunction with this, polyamines might directly affect PrrA itself, and not just the state of the DNA, either altering its conformation or its assembly state.
Based on additional data reported here, which implicates DNA topology as a possible effector of PrrA-regulated gene expression (see below), we favor a model in which the spermidine-dependent increased affinity of P-PrrA binding for RSP3361 gene PrrA site 2, and possibly other PrrA binding DNA sequences, could be due to a favorable change in DNA conformation of the PrrA binding site(s) being promoted by the polyamine, although the above-mentioned possible additional effects cannot be discounted. A substantial number of publications implicate polyamines to affect the topology of DNA; specifically, polyamines can alter DNA bending (31, 68), as well as promote the B-DNA structure to both A-DNA and Z-DNA transitions (32, 80), and affect topoisomerase function (46), for example.
The alluded-to topological effector of gene expression we have examined here is DNA supercoiling. DNA topology, as dictated by supercoiling levels, has been previously reported to regulate the expression of photosynthesis genes, which are positively regulated by PrrA (29, 45) in both R. sphaeroides and R. capsulatus (53, 90). Here, we have developed an assay to show that the expression of pucB (RSP0314 gene), which is regulated by PrrA (28), is also positively regulated by negative supercoiling, using TDTA to change local superhelicity of target promoters in situ, as predicted by the twin-domain model (54). In addition to pucB, the expression of the RSP3361 gene was shown by TDTA to be positively regulated by negative supercoiling, and this was confirmed using the DNA gyrase inhibitor Nv. We favor a model in which P-PrrA binds with higher affinity to RSP3361 gene PrrA site 2 when in a supercoiled topological conformation than when in a linear form. Similar topologically dependent, preferential DNA binding of transcriptional factors has been previously reported for FIS (64) and IHF (75) in E. coli. While DNA supercoiling levels might affect P-PrrA binding to PrrA site 2, an analysis of the RSP3361 gene regulatory region identified an 8-nt sequence, partly overlapping the 3′ end of the −10 region of the presumed promoter and immediate downstream sequence, with a high tendency to become single stranded by absorbing supercoiling energy, as described previously (17, 34). When taken together, these observations prompt us to suggest that negative DNA supercoiling might affect the expression of the RSP3361 gene both by increasing the affinity of P-PrrA binding to PrrA site 2 as well as presumably by promoting the transition from the closed to the open promoter complex to initiate transcription of the RSP3361 gene.
In addition to DNA supercoiling, the intrinsic DNA curvature is another DNA topological determinant with regulatory functions suggested to influence gene expression and possibly PrrA binding to DNA, in R. sphaeroides, either as intrinsic curvature (48, 71) or as DNA bending promoted by DNA binding proteins, like IHF (52). In the case of the RSP3361 gene, neither possibility appears likely, since the PrrA site 2, in the RSP3361 gene, is not predicted to form an intrinsic bend (83), according to our bioinformatics promoter region analysis, and since the expression of the RSP3361 gene seems to be strictly dependent on PrrA (29).
In an accompanying manuscript (28a), we show P-PrrA binding in vitro to genomic DNA sequences present in the regulatory regions of genes positively regulated by PrrA, and containing spacer regions with variable lengths (5 and 8 nucleotides), as predicted previously (58). The orientation of the PrrA half-sites with respect to the longitudinal axis of the DNA helix within DNA sequences containing such different spacing lengths would be predicted to be very different, and additional effectors might be required for optimal gene expression. Thus, we propose that the physiological relevance for DNA supercoiling and polyamines as “regulators” of the expression of the RSP3361 gene, and possibly other genes, might lie in the fact that, in addition to PrrA phosphorylation, which would occur under anaerobic growth conditions, the DNA sites to which this response regulator binds might require a specific topological conformation, as might be the case for other transcriptional regulators (64, 75). This conformation could be dictated by either the length of the spacer region between their PrrA half-sites and/or the DNA sequence of the specific site (28a). In this model, not only the DNA sequence of the PrrA binding site but, also, its topological state would be important determinants to stimulate, or preclude, PrrA binding.
This extra level of regulation would make the PrrA regulon in R. sphaeroides more responsive to additional cellular signals, since the phosphorylation state of PrrA is controlled through redox flow through the electron transport chain (56), and DNA topology might, among other possibilities, (i) be dictated by the terminal electron acceptor used during respiration, as suggested for E. coli (40) and Salmonella enterica (86); (ii) be regulated by intracellular redox conditions, as suggested by the regulation of DNA gyrase activity by thioredoxins in both R. sphaeroides and R. capsulatus (53); and/or (iii) might be an indicator of the physiological state of the cells and influence gene expression, as proposed for intracellular virulence gene expression in S. enterica (13; as reviewed in reference 15).
Acknowledgments
We wish to thank Xiaohua Zeng and Jung Hyeob for helpful discussions and for communicating unpublished data and Stephen Winans for providing plasmid p-Bend-3. We also thank our anonymous reviewers for reading the manuscript critically.
This work was supported by grant GM15590 from the USPHS to S.K.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Arai, H., J. H. Roh, and S. Kaplan. 2008. Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 190286-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachrach, U., Y. C. Wang, and A. Tabib. 2001. Polyamines: new cues in cellular signal transduction. News Physiol. Sci. 16106-109. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, C., S. Elsen, L. R. Swem, D. L. Swem, and S. Masuda. 2003. Redox and light regulation of gene expression in photosynthetic prokaryotes. Philos. Trans. R. Soc. Lond. B 358147-153. (Discussion, 358:153-154.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, T. H., S. Du, and C. E. Bauer. 1999. Autophosphorylation, phosphotransfer, and DNA-binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J. Biol. Chem. 27416343-16348. [DOI] [PubMed] [Google Scholar]
- 5.Braatsch, S., O. V. Moskvin, G. Klug, and M. Gomelsky. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J. Bacteriol. 1867726-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruscella, P., J. M. Eraso, J. H. Roh, and S. Kaplan. 2008. The use of chromatin immunoprecipitation to define PpsR binding activity in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1906817-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buratowski, S., and L. A. Chodosh. 2001. Mobility shift DNA-binding assay using gel electrophoresis. Curr. Protoc. Mol. Biol. 1212.2. [DOI] [PubMed] [Google Scholar]
- 8.Callister, S. J., M. A. Dominguez, C. D. Nicora, X. Zeng, C. L. Tavano, S. Kaplan, T. J. Donohue, R. D. Smith, and M. S. Lipton. 2006. Application of the accurate mass and time tag approach to the proteome analysis of sub-cellular fractions obtained from Rhodobacter sphaeroides 2.4.1. Aerobic and photosynthetic cell cultures. J. Proteome Res. 51940-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callister, S. J., C. D. Nicora, X. Zeng, J. H. Roh, M. A. Dominguez, C. L. Tavano, M. E. Monroe, S. Kaplan, T. J. Donohue, R. D. Smith, and M. S. Lipton. 2006. Comparison of aerobic and photosynthetic Rhodobacter sphaeroides 2.4.1 proteomes. J. Microbiol. Methods 67424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, S. S. 1988. A guide to polyamines. Oxford University Press, New York, NY.
- 11.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell Physiol. 4925-68. [DOI] [PubMed] [Google Scholar]
- 12.Comolli, J. C., A. J. Carl, C. Hall, and T. Donohue. 2002. Transcriptional activation of the Rhodobacter sphaeroides cytochrome c2 gene P2 promoter by the response regulator PrrA. J. Bacteriol. 184390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cróinín, O. T., R. K. Carroll, A. Kelly, and C. J. Dorman. 2006. Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 62869-882. [DOI] [PubMed] [Google Scholar]
- 14.Davis, J., T. J. Donohue, and S. Kaplan. 1988. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J. Bacteriol. 170320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorman, C. J., and C. P. Corcoran. 2009. Bacterial DNA topology and infectious disease. Nucleic Acids Res. 37672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dow, L. K., A. Changela, H. E. Hefner, and M. E. Churchill. 1997. Oxidation of a critical methionine modulates DNA binding of the Drosophila melanogaster high mobility group protein, HMG-D. FEBS Lett. 414514-520. [DOI] [PubMed] [Google Scholar]
- 17.Drew, H. R., J. R. Weeks, and A. A. Travers. 1985. Negative supercoiling induces spontaneous unwinding of a bacterial promoter. EMBO J. 41025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dryden, S. C., and S. Kaplan. 1993. Identification of cis-acting regulatory regions upstream of the rRNA operons of Rhodobacter sphaeroides. J. Bacteriol. 1756392-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du, S., T. H. Bird, and C. E. Bauer. 1998. DNA binding characteristics of RegA. A constitutively active anaerobic activator of photosynthesis gene expression in Rhodobacter capsulatus. J. Biol. Chem. 27318509-18513. [DOI] [PubMed] [Google Scholar]
- 20.Dubbs, J. M., and F. R. Tabita. 2003. Interactions of the cbbII promoter-operator region with CbbR and RegA (PrrA) regulators indicate distinct mechanisms to control expression of the two cbb operons of Rhodobacter sphaeroides. J. Biol. Chem. 27816443-16450. [DOI] [PubMed] [Google Scholar]
- 21.Dubbs, J. M., and F. R. Tabita. 2004. Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28353-376. [DOI] [PubMed] [Google Scholar]
- 22.Elsen, S., M. Jaubert, D. Pignol, and E. Giraud. 2005. PpsR: a multifaceted regulator of photosynthesis gene expression in purple bacteria. Mol. Microbiol. 5717-26. [DOI] [PubMed] [Google Scholar]
- 23.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emmerich, R., P. Strehler, H. Hennecke, and H. M. Fischer. 2000. An imperfect inverted repeat is critical for DNA binding of the response regulator RegR of Bradyrhizobium japonicum. Nucleic Acids Res. 284166-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eraso, J. M., and S. Kaplan. 1996. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1787037-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eraso, J. M., and S. Kaplan. 2000. From redox flow to gene regulation: role of the PrrC protein of Rhodobacter sphaeroides 2.4.1. Biochemistry 392052-2062. [DOI] [PubMed] [Google Scholar]
- 27.Eraso, J. M., and S. Kaplan. 1995. Oxygen-insensitive synthesis of the photosynthetic membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J. Bacteriol. 1772695-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eraso, J. M., and S. Kaplan. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 17632-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Eraso, J. M., and S. Kaplan. 2009. Half-site DNA sequence of PrrA site 2 of RSP3361. J. Bacteriol. 1914353-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eraso, J. M., J. H. Roh, X. Zeng, S. J. Callister, M. S. Lipton, and S. Kaplan. 2008. Role of the transcriptional regulator PrrA in Rhodobacter sphaeroides 2.4.1: combined transcriptome and proteome analysis. J. Bacteriol. 1904831-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farwer, J., M. J. Packer, and C. A. Hunter. 2007. PREDICTOR: a web-based tool for the prediction of atomic structure from sequence for double helical DNA with up to 150 base pairs. In Silico Biol. 70052. [PubMed] [Google Scholar]
- 31.Feuerstein, B. G., N. Pattabiraman, and L. J. Marton. 1990. Molecular mechanics of the interactions of spermine with DNA: DNA bending as a result of ligand binding. Nucleic Acids Res. 181271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feuerstein, B. G., L. D. Williams, H. S. Basu, and L. J. Marton. 1991. Implications and concepts of polyamine-nucleic acid interactions. J. Cell Biochem. 4637-47. [DOI] [PubMed] [Google Scholar]
- 33.Fradet-Turcotte, A., C. Vincent, S. Joubert, P. A. Bullock, and J. Archambault. 2007. Quantitative analysis of the binding of simian virus 40 large T antigen to DNA. J. Virol. 819162-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fye, R. M., and C. J. Benham. 1999. Exact method for numerically analyzing a model of local denaturation in superhelically stressed DNA. Phys. Rev. E 592408-3426. [Google Scholar]
- 35.Gomelsky, M., and S. Kaplan. 1995. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1774609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomelsky, M., and S. Kaplan. 1995. Isolation of regulatory mutants in photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1 and partial complementation of a PrrB mutant by the HupT histidine-kinase. Microbiology 141(Pt. 8)1805-1819. [DOI] [PubMed] [Google Scholar]
- 37.Hamana, K., M. Kamekura, H. Onishi, T. Akazawa, and S. Matsuzaki. 1985. Polyamines in photosynthetic eubacteria and extreme-halophilic archaebacteria. J. Biochem. 971653-1658. [DOI] [PubMed] [Google Scholar]
- 38.Hemschemeier, S. K., U. Ebel, A. Jager, A. Balzer, M. Kirndorfer, and G. Klug. 2000. In vivo and in vitro analysis of RegA response regulator mutants of Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 2291-300. [PubMed] [Google Scholar]
- 39.Hemschemeier, S. K., M. Kirndorfer, M. Hebermehl, and G. Klug. 2000. DNA binding of wild type RegA protein and its differential effect on the expression of pigment binding proteins in Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 2235-243. [PubMed] [Google Scholar]
- 40.Hsieh, L. S., R. M. Burger, and K. Drlica. 1991. Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J. Mol. Biol. 219443-450. [DOI] [PubMed] [Google Scholar]
- 41.Igarashi, K., and K. Kashiwagi. 1999. Polyamine transport in bacteria and yeast. Biochem. J. 344(Pt. 3)633-642. [PMC free article] [PubMed] [Google Scholar]
- 42.Jones, D. F., R. A. Stenzel, and T. J. Donohue. 2005. Mutational analysis of the C-terminal domain of the Rhodobacter sphaeroides response regulator PrrA. Microbiology 1514103-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung, I. L., and I. G. Kim. 2003. Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem. Biophys. Res. Commun. 301915-922. [DOI] [PubMed] [Google Scholar]
- 44.Kang, Y., K. D. Weber, Y. Qiu, P. J. Kiley, and F. R. Blattner. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 1871135-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaplan, S., J. Eraso, and J. H. Roh. 2005. Interacting regulatory networks in the facultative photosynthetic bacterium, Rhodobacter sphaeroides 2.4.1. Biochem. Soc. Trans. 3351-55. [DOI] [PubMed] [Google Scholar]
- 46.Krasnow, M. A., and N. R. Cozzarelli. 1982. Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J. Biol. Chem. 2572687-2693. [PubMed] [Google Scholar]
- 47.Labes, M., A. Puhler, and R. Simon. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene 8937-46. [DOI] [PubMed] [Google Scholar]
- 48.Laguri, C., M. K. Phillips-Jones, and M. P. Williamson. 2003. Solution structure and DNA binding of the effector domain from the global regulator PrrA (RegA) from Rhodobacter sphaeroides: insights into DNA binding specificity. Nucleic Acids Res. 316778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laguri, C., R. A. Stenzel, T. J. Donohue, M. K. Phillips-Jones, and M. P. Williamson. 2006. Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides. Biochemistry 457872-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, J. K., and S. Kaplan. 1992. cis-Acting regulatory elements involved in oxygen and light control of puc operon transcription in Rhodobacter sphaeroides. J. Bacteriol. 1741146-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee, J. K., and S. Kaplan. 1992. Isolation and characterization of trans-acting mutations involved in oxygen regulation of puc operon transcription in Rhodobacter sphaeroides. J. Bacteriol. 1741158-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee, J. K., S. Wang, J. M. Eraso, J. Gardner, and S. Kaplan. 1993. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. Involvement of an integration host factor-binding sequence. J. Biol. Chem. 26824491-24497. [PubMed] [Google Scholar]
- 53.Li, K., C. Pasternak, E. Hartig, K. Haberzettl, A. Maxwell, and G. Klug. 2004. Thioredoxin can influence gene expression by affecting gyrase activity. Nucleic Acids Res. 324563-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 847024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, Z., and J. W. Little. 1998. The spacing between binding sites controls the mode of cooperative DNA-protein interactions: implications for evolution of regulatory circuitry. J. Mol. Biol. 278331-338. [DOI] [PubMed] [Google Scholar]
- 56.Mackenzie, C., J. M. Eraso, M. Choudhary, J. H. Roh, X. Zeng, P. Bruscella, A. Puskas, and S. Kaplan. 2007. Postgenomic adventures with Rhodobacter sphaeroides. Annu. Rev. Microbiol. 61283-307. [DOI] [PubMed] [Google Scholar]
- 57.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 58.Mao, L., C. Mackenzie, J. H. Roh, J. M. Eraso, S. Kaplan, and H. Resat. 2005. Combining microarray and genomic data to predict DNA binding motifs. Microbiology 1513197-3213. [DOI] [PubMed] [Google Scholar]
- 59.McClure, W. R., D. K. Hawley, P. Youderian, and M. M. Susskind. 1983. DNA determinants of promoter selectivity in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 47(Pt. 1)477-481. [DOI] [PubMed] [Google Scholar]
- 60.Meinhardt, S. W., P. J. Kiley, S. Kaplan, A. R. Crofts, and S. Harayama. 1985. Characterization of light-harvesting mutants of Rhodopseudomonas sphaeroides. I. Measurement of the efficiency of energy transfer from light-harvesting complexes to the reaction center. Arch. Biochem. Biophys. 236130-139. [DOI] [PubMed] [Google Scholar]
- 61.Moskvin, O. V., L. Gomelsky, and M. Gomelsky. 2005. Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as a master regulator of photosystem development. J. Bacteriol. 1872148-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh, J. I., and S. Kaplan. 2002. Oxygen adaptation. The role of the CcoQ subunit of the cbb3 cytochrome c oxidase of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 27716220-16228. [DOI] [PubMed] [Google Scholar]
- 63.Oh, J. I., I. J. Ko, and S. Kaplan. 2004. Reconstitution of the Rhodobacter sphaeroides cbb3-PrrBA signal transduction pathway in vitro. Biochemistry 437915-7923. [DOI] [PubMed] [Google Scholar]
- 64.Opel, M. L., K. A. Aeling, W. M. Holmes, R. C. Johnson, C. J. Benham, and G. W. Hatfield. 2004. Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism. Mol. Microbiol. 53665-674. [DOI] [PubMed] [Google Scholar]
- 65.Opel, M. L., and G. W. Hatfield. 2001. DNA supercoiling-dependent transcriptional coupling between the divergently transcribed promoters of the ilvYC operon of Escherichia coli is proportional to promoter strengths and transcript lengths. Mol. Microbiol. 39191-198. [DOI] [PubMed] [Google Scholar]
- 66.Panagiotidis, C. A., S. Artandi, K. Calame, and S. J. Silverstein. 1995. Polyamines alter sequence-specific DNA-protein interactions. Nucleic Acids Res. 231800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 1864748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pastré, D., O. Pietrement, F. Landousy, L. Hamon, I. Sorel, M. O. David, E. Delain, A. Zozime, and E. Le Cam. 2006. A new approach to DNA bending by polyamines and its implication in DNA condensation. Eur. Biophys. J. 35214-223. [DOI] [PubMed] [Google Scholar]
- 69.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 1762869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peter, B. J., J. Arsuaga, A. M. Breier, A. B. Khodursky, P. O. Brown, and N. R. Cozzarelli. 2004. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 5R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ranson-Olson, B., D. F. Jones, T. J. Donohue, and J. H. Zeilstra-Ryalls. 2006. In vitro and in vivo analysis of the role of PrrA in Rhodobacter sphaeroides 2.4.1 hemA gene expression. J. Bacteriol. 1883208-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roh, J. H., W. E. Smith, and S. Kaplan. 2004. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1. Redox active gene expression profile. J. Biol. Chem. 2799146-9155. [DOI] [PubMed] [Google Scholar]
- 73.Sabbah, M., S. Le Ricousse, G. Redeuilh, and E. E. Baulieu. 1992. Estrogen receptor-induced bending of the Xenopus vitellogenin A2 gene hormone response element. Biochem. Biophys. Res. Commun. 185944-952. [DOI] [PubMed] [Google Scholar]
- 74.Shah, P., D. G. Romero, and E. Swiatlo. 2008. Role of polyamine transport in Streptococcus pneumoniae response to physiological stress and murine septicemia. Microb. Pathog. 45167-172. [DOI] [PubMed] [Google Scholar]
- 75.Sheridan, S. D., C. J. Benham, and G. W. Hatfield. 1998. Activation of gene expression by a novel DNA structural transmission mechanism that requires supercoiling-induced DNA duplex destabilization in an upstream activating sequence. J. Biol. Chem. 27321298-21308. [DOI] [PubMed] [Google Scholar]
- 76.Swem, L. R., S. Elsen, T. H. Bird, D. L. Swem, H. G. Koch, H. Myllykallio, F. Daldal, and C. E. Bauer. 2001. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol. 309121-138. [DOI] [PubMed] [Google Scholar]
- 77.Swem, L. R., D. L. Swem, J. Wu, and C. E. Bauer. 2007. Purification and assays of Rhodobacter capsulatus RegB-RegA two-component signal transduction system. Methods Enzymol. 422171-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 4981-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tai, T. N., W. A. Havelka, and S. Kaplan. 1988. A broad-host-range vector system for cloning and translational lacZ fusion analysis. Plasmid 19175-188. [DOI] [PubMed] [Google Scholar]
- 80.Thomas, T. J., U. B. Gunnia, and T. Thomas. 1991. Polyamine-induced B-DNA to Z-DNA conformational transition of a plasmid DNA with (dG-dC)n insert. J. Biol. Chem. 2666137-6141. [PubMed] [Google Scholar]
- 81.van Niel, C. B. 1944. The culture, general physiology, and classification of the non-sulfur purple and brown bacteria. Bacteriol. Rev. 81-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vannier-Santos, M. A., D. Menezes, M. F. Oliveira, and F. G. de Mello. 2008. The putrescine analogue 1,4-diamino-2-butanone affects polyamine synthesis, transport, ultrastructure and intracellular survival in Leishmania amazonensis. Microbiology 1543104-3111. [DOI] [PubMed] [Google Scholar]
- 83.Vlahovicek, K., L. Kajan, and S. Pongor. 2003. DNA analysis servers: plot.it, bend.it, model.it and IS. Nucleic Acids Res. 313686-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Willett, J., J. L. Smart, and C. E. Bauer. 2007. RegA control of bacteriochlorophyll and carotenoid synthesis in Rhodobacter capsulatus. J. Bacteriol. 1897765-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wortham, B. W., C. N. Patel, and M. A. Oliveira. 2007. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 603106-115. [DOI] [PubMed] [Google Scholar]
- 86.Yamamoto, N., and M. L. Droffner. 1985. Mechanisms determining aerobic or anaerobic growth in the facultative anaerobe Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 822077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshida, M., K. Kashiwagi, A. Shigemasa, S. Taniguchi, K. Yamamoto, H. Makinoshima, A. Ishihama, and K. Igarashi. 2004. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 27946008-46013. [DOI] [PubMed] [Google Scholar]
- 88.Zeilstra-Ryalls, J., and S. Kaplan. 1998. Role of the fnrL gene in photosystem gene expression and photosynthetic growth of Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1801496-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeng, X., J. H. Roh, S. J. Callister, C. L. Tavano, T. J. Donohue, M. S. Lipton, and S. Kaplan. 2007. Proteomic characterization of the Rhodobacter sphaeroides 2.4.1 photosynthetic membrane: identification of new proteins. J. Bacteriol. 1897464-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu, Y. S., and J. E. Hearst. 1988. Transcription of oxygen-regulated photosynthetic genes requires DNA gyrase in Rhodobacter capsulatus. Proc. Natl. Acad. Sci. USA 854209-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]