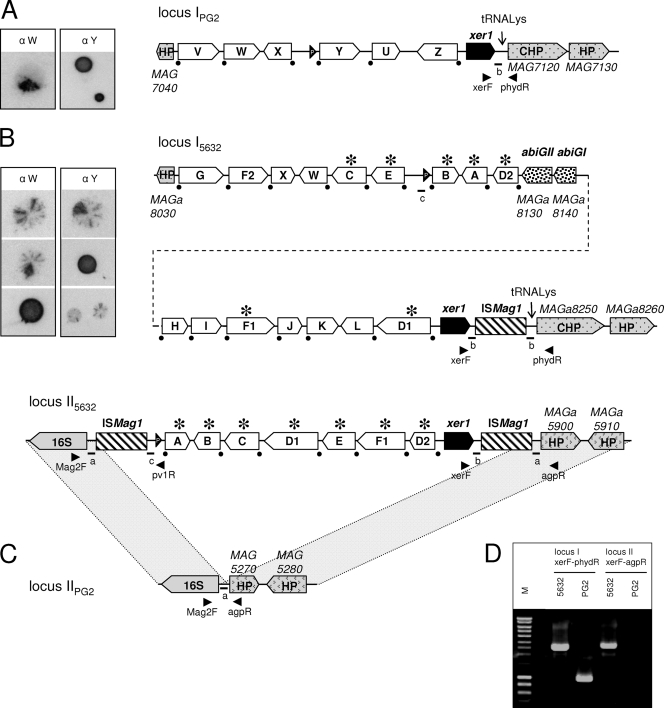
FIG. 1.
Comparison of M. agalactiae vpma loci between the PG2 type strain and strain 5632. Schematics represent the organization of the vpma loci in clonal variant 55.5 derived from PG2 (16, 42) (A) and in clonal variant c1 derived from strain 5632 (B). (C) Counterpart of locus II5632 in PG2 showing the absence of vpma genes in this region. (D) The presence of two distinct loci in 5632 was confirmed by PCR, using the primer pair xerF-phydR or xerF-agpR, and the resulting amplicons are shown. The locations of the primers are indicated by arrowheads in panels A, B, and C. Large white arrows labeled with letters represent Vpma CDSs. The positions of the promoters are represented by black arrowheads labeled “P.” The two non-Vpma-related CDSs (abiGI and abiGII) are indicated by large arrows filled with a dotted pattern. ISMag1 elements are indicated by hatched boxes. Recombination sites downstream of each vpma gene are indicated by black dots. An asterisk indicates that the corresponding vpma gene is present at two distinct loci. Schematics were drawn approximately to scale. HP, hypothetical protein; CHP, conserved hypothetical protein. Small letters and bars indicate the positions of short particular sequences mentioned in the text and in Fig. 3 and 4. The pictures on the left side of panels A and B illustrate the variable surface expression of Vpma, as previously described (8, 17). These correspond to colony immunoblots using Vpma-specific polyclonal antibodies recognizing PG2 VpmaW (α W) and VpmaY (α Y) epitopes.