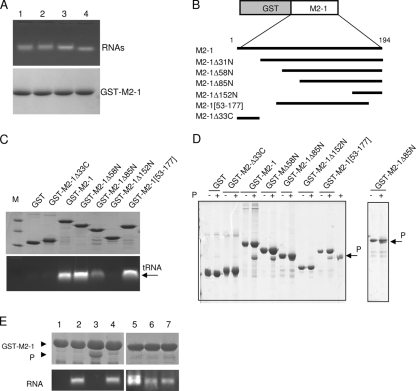
FIG. 7.
The domain of residues 59 to 177 is sufficient for binding to P and RNA in a competitive manner. (A) Binding of different RNAs to GST-M2-1. GST-M2-1 complexes bound to glutathione-Sepharose beads (100 μg) were incubated with 100 μg of RNA transcribed from pCDNA3 (lane 1), pET-P (lane 2), or a leader-NS1 sequence (lane 3) or 100 μg of yeast tRNA (lane 4) for 1 h at room temperature. After extensive washing, GST-M2-1-RNA complexes were analyzed by agarose gel electrophoresis by double staining with EtBr (top) and amido black (bottom) to reveal RNAs and proteins, respectively. (B) Schematic representation of the deletion mutants of the GST-M2-1 fusion protein that were used in the pulldown assays. (C and D) Mapping of the RNA (C)- and P (D)-binding domains on M2-1 by in vitro pulldown assays using deletion mutants of the GST-M2-1 fusion protein, tRNA, and purified recombinant RSV P protein. After extensive washing, the sample of GST-M2-1-RNA complexes was separated into two equal parts and analyzed by agarose gel electrophoresis and EtBr staining (C, bottom) and SDS-PAGE and Coomassie blue staining (C, top) to reveal RNA and proteins, respectively. Lane M, molecular size markers; +, M2-1 was inoculated with P in vitro. (E) Evidence for competition between RNA and P for binding to M2-1. Glutathione-Sepharose beads containing RNA-free GST-M2-1 (lane 1) were incubated with tRNA alone (lane 2), P alone (lane 3), or P in the presence of yeast tRNA (lane 4), an 81-nucleotide RNA transcribed from pBluescript plasmid (lane 5), or an 80-nucleotide RNA containing the HRSV leader sequence in the plus (lane 6) or minus (lane 7) polarity. After further incubation for 1 h at 20°C, the reaction products were washed extensively and separated into two parts for analysis to detect the presence of both P (top) and RNA (bottom) bound to M2-1, as described above. The GST-M2-1 and P protein bands are indicated by arrowheads.