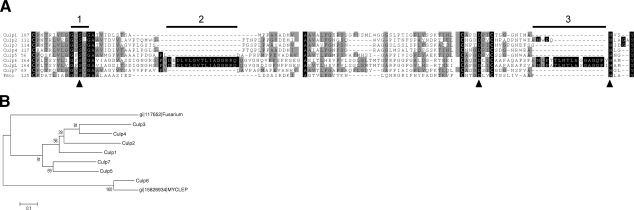
Figure 1.
Alignment and phylogram of the CULPs of M. tuberculosis, M. leprae, and F. solani. A) Alignment of residues of the catalytic triad (indicated by arrowheads) and surrounding regions. Residues conserved in all proteins are indicated with black background; those shaded gray are identical in ≥50% of the proteins. Conserved cysteines are boldface in black background. Bar 1 indicates the conserved pentapeptide motif G-X-S-X-G containing the catalytic serine. Bars 2 and 3 indicate insertion of amino acids found in Culp6 and in the M. leprae ortholog, respectively. B) Bootstrapped phylogram from alignments of whole sequences. Numbers represent percentage bootstrap support. Scale bar indicates 10% amino acid sequence divergence. Symbol gi| indicates the NCBI accession number for the F. solani and M. leprae (MYCLEP) proteins.