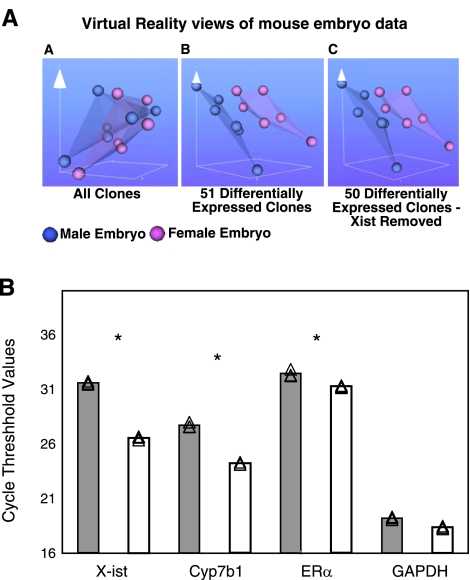
Figure 4.
Sex differences in gene expression in unstressed ED10.5 whole embryos. A) Virtual reality views of paired male/female embryo microarray intensity data created using the BioMiner VT module. Each sphere represents intensity data from one channel of paired male/female microarray. Blue spheres represent male data, pink represent female data. VR representation of all clones (AA), 51 differentially expressed clones (AB), and the 50 differentially expressed clones with the clone for X-ist removed (AC). B) qRT-PCR verification of sample genes from the microarray. Source of cDNA was whole male or female embryo homogenate at ED10.5. Samples were normalized at the RNA and cDNA levels for equal cDNA loading. Gray bars, male; white bars, female. Experiments were done in triplicate. Individual CT points are shown together with mean values. *P < 0.05; Student’s t test. Ordinate is average CT of fluorescence detection for real-time PCR. Each increment in CT indicates a starting transcript concentration ∼0.5× that of CT-1. X-ist was expressed significantly more in female embryos, while ERα and Cyp7b1 were higher, and GAPDH was not sexually dimorphic.