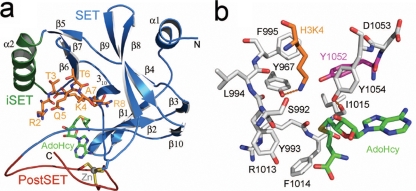
FIG. 1.
Homology model of the yeast Set1 catalytic domain. (a) Ribbon representation of the secondary structure of the Set1 catalytic domain illustrating the SET domain (blue), the inserted SET motif (iSET, green), and the PostSET domain (red). A histone H3 substrate peptide spanning residues 2 to 8 (orange carbon atoms) and the product, AdoHcy (green carbons), are docked in the active site. The side chains of the four cysteines that coordinate the Zn(II) ion in the PostSET domain are also illustrated. (b) Active site of Set1. Residues comprising the enzyme's lysine binding channel are shown with gray carbon atoms, with the exception of the Phe/Tyr switch residue Tyr1052 that is highlighted by magenta carbons. AdoHcy and H3K4 are depicted as described for panel A. Hydrogen bonds are illustrated by orange dashed lines.