Table 3.
Number of minimal absent words and generic absent words for some genomes.
| Organism | Reference | Genome size | 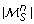 |
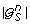 |
Length, n |
| 104 | 104 | 11 | |||
| H. sapiens | Release 36.1 | ≈ 2.9 Gb | 44 149 | 44 970 | 12 |
| 2 039 862 | 2 368 682 | 13 | |||
| 190 | 190 | 11 | |||
| M. musculus | Release m36.1 | ≈ 2.6 Gb | 52 087 | 53 573 | 12 |
| 2 192 708 | 2 579 838 | 13 | |||
| 104 | 104 | 11 | |||
| D. melanogaster | FB 5 | ≈ 162 Mb | 172 849 | 173 674 | 12 |
| 10 040 282 | 11 335 034 | 13 | |||
| 2 | 2 | 10 | |||
| C. elegans | WB 170 | ≈ 100 Mb | 7 664 | 7 680 | 11 |
| 1 092 286 | 1 151 728 | 12 | |||
| 2 262 | 2 262 | 11 | |||
| N. crassa | Assembly 7 | ≈ 39 Mb | 1 064 938 | 1 082 787 | 12 |
| 20 213 298 | 27 903 272 | 13 | |||
| 2 | 2 | 9 | |||
| S. cerevisiae S228C | SGD 1 | ≈ 12 Mb | 6 435 | 6 450 | 10 |
| 414 520 | 462 882 | 11 | |||
| 248 | 248 | 8 | |||
| S. aureus MSSA476 | NC002953 | ≈ 2.8 Mb | 11 908 | 13 744 | 9 |
| 162 113 | 251 497 | 10 | |||
| 1 | 1 | 8 | |||
| T. kodakarensis | NC006624 | ≈ 2.09 Mb | 2 314 | 2 322 | 9 |
| 136 917 | 154 340 | 10 | |||
| 3 | 3 | 6 | |||
| M. jannaschii | NC000909 | ≈ 1.66 Mb | 126 | 150 | 7 |
| 3 790 | 4 834 | 8 | |||
| 5 | 5 | 6 | |||
| M. genitalium | NC000908 | ≈ 0.58 Mb | 340 | 380 | 7 |
| 6 156 | 8 733 | 8 | |||
The notation ![]() corresponds to the number of minimal absent words of length n associated with string S, whereas
corresponds to the number of minimal absent words of length n associated with string S, whereas ![]() has a similar meaning but for the case of generic absent words. The generic absent words have been generated using publicly available software provided by Herold et al. 3. The organisms are sorted according to decreasing genome size, which refers to the number of unambiguous bases of the genome. The reversed complement of the sequences has been considered in the generation of the results.
has a similar meaning but for the case of generic absent words. The generic absent words have been generated using publicly available software provided by Herold et al. 3. The organisms are sorted according to decreasing genome size, which refers to the number of unambiguous bases of the genome. The reversed complement of the sequences has been considered in the generation of the results.
