Abstract
Background
The Notch signaling pathway is an evolutionary conserved signal transduction pathway involved in embryonic patterning and regulation of cell fates during development and self-renewal. Recent studies have demonstrated that this pathway is integral to a complex system of interactions, involving as well other signal transduction pathways, and implicated in distinct human diseases. Delta-like 1 (Dll1) is one of the known ligands of the Notch receptors. The role of the Notch ligands is less well understood. Loss-of-function of Dll1 leads to embryonic lethality, but reduction of Delta-like 1 protein levels has not been studied in adult stage.
Methodology/Principal Findings
Here we present the haploinsufficient phenotype of Dll1 and a missense mutant Dll1 allele (Dll1C413Y). Haploinsufficiency leads to a complex phenotype with several biological processes altered. These alterations reveal the importance of Dll1 mainly in metabolism, energy balance and in immunology. The animals are smaller, lighter, with altered fat to lean ratio and have increased blood pressure and a slight bradycardia. The animals have reduced cholesterol and triglyceride levels in blood. At the immunological level a subtle phenotype is observed due to the effect and fine-tuning of the signaling network at the different levels of differentiation, proliferation and function of lymphocytes. Moreover, the importance of the proteolytic regulation of the Notch signaling network emphasized.
Conclusions/Significance
In conclusion, slight alterations in one player of Notch signaling alter the entire organism, emphasizing the fine-tuning character of this pathway in a high number of processes.
Introduction
The Notch signaling pathway is an intercellular signaling mechanism that is highly conserved during evolution of vertebrates. It is involved in the determination of cell fates in different cell types during embryonic development. First described in the fly, Notch orthologs have been identified successively in many species including birds, rodents and man. In mammals four distinct Notch genes (Notch1–Notch4) have been identified. The Notch receptors are anchored to the membrane via a single transmembrane domain. Two families of transmembrane ligands interact with the Notch receptors, the Delta and the Jagged/Serrate proteins. In mammals three Delta and two Jagged/Serrate class proteins have been described. The canonical Notch signaling pathway involves proteolytic processing events that cleave the Notch receptors in response to ligand activation, leading to the activation of target genes (frequently basic helix-loop-helix transcriptional factors) (for review see [1]–[3]).
Loss- or gain-of-function of the Notch receptors or their ligands have diverse and often severe clinical, physiological and biological consequences in humans (for review see [4], [5]). T-cell acute lymphoblastic leukemia, aortic valve disease, and familial forms of cardiomyopathy, have been associated with mutations in Notch1. Alagille syndrome, a developmental disease, is associated with mutations in Jagged1 and in less extent with mutations in Notch2. Mutations in Notch3 lead to a neurological disease, a cerebral arteriopathy autosomal dominant disease with subcortical infarcts and leukoencephalopathy abbreviated as CADASIL. Mutations in Delta3 have been associated with spondylocostal dysostosis which leads to severe congenital malformations of the vertebral column. Notch signaling and associated disease involve not only these inherited disorders. Recent publications revealed the involvement of Notch signaling in tumor associated angiogenesis (for review see [6], [7]). Experiments in animal models show that the Notch signaling pathway also plays an important role in tissue regeneration after injury of adult organs, such as heart, liver, brain, kidney and pancreas [8]–[12]. Therefore, Notch signaling has many diverse roles in physiology and pathology.
The fine-tuning of this complex system of regulatory interactions with pleiotropic functions is essential to avoid disease states, but not yet fully understood. One efficient approach to identify components of a molecular network is to perform genetic modifier- or sensitized-screens [13]. Recently, we reported the identification of phenotypic modifiers in a sensitized screen using Dll1 heterozygous (Dll1tm1Gos/+) animals [14]. A complementary approach to better understand pleiotropic Notch signaling functions is to investigate the haploinsufficient phenotype of Notch pathway members, since most homozygous Notch mutants and mutant alleles of interacting proteins or ligands are embryonic lethal [15]–[19]. Here, we have comprehensively analyzed the phenotype of Dll1tm1Gos heterozygous animals and of a missense mutant Dll1 allele (Dll1C413Y) in 14 phenotyping platforms covering more than 350 parameters at the German Mouse Clinic (GMC) [20]–[22].
Haploinsufficiency was recently investigated for the Notch ligand Jagged1 in two inbred mouse strains with distinct phenotypes [23], so the genetic background of the inbred mouse strain may affect the observable mutant phenotype. We present here the haploinsufficient phenotype of Dll1tm1Gos/+ mice on the C3HeB/FeJ genetic background and compare the results to previously recorded data on the 129X1/SvJ genetic background.
Results
Genetic background of Dll1tm1Gos/+ mice
We outcrossed the mice carrying the Dll1tm1Gos allele [15] from a 129X1/SvJ (129X1) genetic background for 11 generations to the C3HeB/FeJ (C3H) genetic background. The 129X1 genetic background was recently identified as being non-isogenic [14]. In the C3.Dll1tm1Gos/+ animals only one of 137 genome-wide SNPs showed heterozygosity (data not shown). This SNP (rs13482869) is at a distance of approximately 5 Mb to the Dll1 gene in which the lacZ reporter gene was inserted to disrupt the Dll1 coding sequence [15].
Body weight, size and body composition
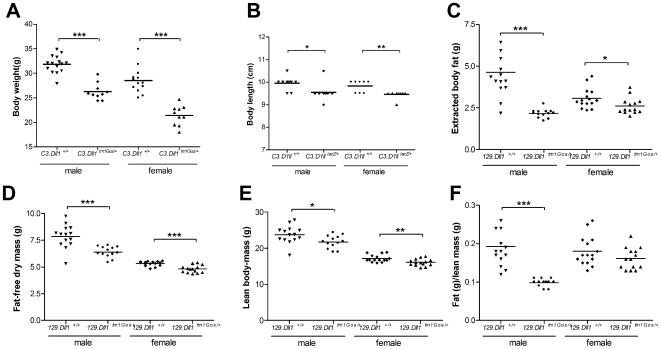
C3.Dll1tm1Gos/+ animals are significantly lighter than wild-type littermates (Figure 1A). This difference in body weight is more pronounced in C3H than in 129X1 and mixed (129X1/SvJ.C57BL6/J) genetic backgrounds [14]. Heterozygous C3H females showed on average an 18% to 20% reduction and heterozygous C3H males on average an 11% to 15% reduction in their body weight at 15 weeks after birth (depending on the analyzed cohort, see Figure 1A and Table S1). The average body weights of C3.Dll1tm1Gos/+ mice were also reduced at 6, 8, 10, and 12 weeks of age (Table S1).
Figure 1. Body weight, size and composition in heterozygous Dll1tm1Gos/+ animals compared to wild-type littermates.
A–B) Body weight and body length were determined at 15 and 16 weeks of age, respectively, in two different cohorts. Depicted are the scatter dot plot representations and the mean for each group: heterozygous C3.Dll1tm1Gos/+ males (weight, n = 13; length, n = 9) and their wild-type littermates (C3.Dll1+/+, weight, n = 15; length, n = 10) and heterozygous C3.Dll1tm1Gos/+ females (n = 10) and their wild-type littermates (C3.Dll1+/+, weight, n = 10; length, n = 9). C–F) Selected metabolic parameters were recorded using the Soxhlet lipid extraction with petrol ether as solvent in the tertiary screen at the GMC at 30–32 weeks of age. Depicted are the scatter dot plot representations for and the mean for each group: heterozygous 129.Dll1tm1Gos/+ males (n = 13) and their wild-type littermates (C3.Dll1+/+, n = 14); heterozygous C3.Dll1tm1Gos/+ females (n = 14) and their wild-type littermates (129.Dll1+/+, n = 15). P-value calculated performing unpaired t-test when variances were not significantly different. If significantly different then the Mann-Whitney test was performed. P-value: * P<0.05, ** P<0.01, *** P<0.001.
Male and female Dll1tm1Gos/+ animals were significantly shorter than their wild-type littermates (Figure 1B). To investigate body composition a cohort of mutant and wild-type 129X1 mice entered the metabolic screen at 30 to 32 weeks of age. No significant differences in the water contents of wild-type and Dll1tm1Gos/+ animals could be detected (data not shown). Using the Soxhlet lipid extraction with petrol ether as solvent the body fat, fat-free dry mass, and lean-body mass of the mice were measured. 129.Dll1tm1Gos/+ animals showed a significantly reduced average body fat content (Figure 1C), reduced average fat-free dry mass (Figure 1D) and a reduced average lean-body mass (Figure 1E) when compared to wild-type littermates. Importantly, the average fat to lean mass ratio was significantly reduced in male and slightly reduced in female heterozygous mutants when compared to wild-type animals (Figure 1F). Similarly, C3.Dll1tm1Gos/+ animals showed reduced body fat and lean mass based upon DEXA densitometry (see Table S1).
In summary, Dll1tm1Gos/+ animals are shorter; show a reduced body weight and an altered body composition, independently from the two genetic backgrounds.
Energy metabolism
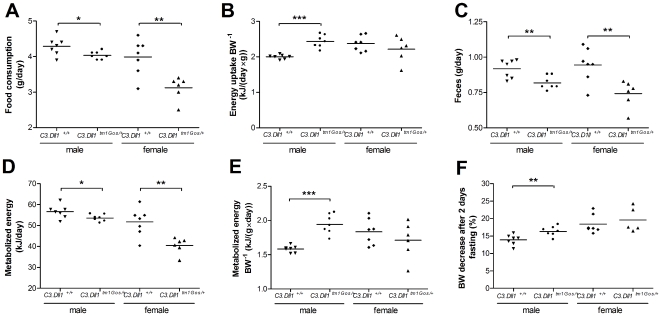
C3.Dll1tm1Gos/+ animals on average consumed significantly less food ad libitum and had a reduced energy uptake as compared to wild-type littermates (Figure 2A). However, when energy uptake was normalized to body weight only the male Dll1tm1Gos/+ mice showed a significant increase of energy uptake in comparison to wild-type males. No such difference in body weight normalized energy uptake was observed between wild-type and C3.Dll1tm1Gos/+ females (Figure 2B).
Figure 2. Metabolic parameters collected from heterozygous C3.Dll1tm1Gos/+ animals and their wild-type littermates at the GMC.
A–E) Values were recorded during 2 weeks ad libitum feeding. F) Body weight decrease (%) after two days fasting. Depicted are the scatter dot plot representations and the mean for each group: heterozygous C3.Dll1tm1Gos/+ males (n = 7) and their wild-type littermates (C3.Dll1+/+, n = 7); heterozygous C3.Dll1tm1Gos/+ females (n = 6) and their wild-type littermates (C3.Dll1+/+, n = 7). P-value calculated performing unpaired t-test when variances were not significantly different. If significantly different then the Mann-Whitney test was performed. P-value: * P<0.05, ** P<0.01, *** P<0.001.
As a consequence of decreased food intake, both male and female C3.Dll1tm1Gos/+ animals showed significantly reduced feces excretion per day compared to wild-type littermates (Figure 2C). The metabolized energy of individual mice was measured by subtracting the energy content of collected feces and urine from the energy uptake. Both female and male C3.Dll1tm1Gos/+ mice showed significantly reduced metabolized energy (Figure 2D). When normalized to the individual body weights, significantly increased metabolized energy was detected only for the group of C3.Dll1tm1Gos/+ males (Figure 2E). In contrast, in 129.Dll1tm1Gos/+ mice only females showed increased energy uptake and metabolized energy compared to wild-type female littermates. The tendency towards this difference in both parameters was also evident for male 129.Dll1tm1Gos/+ mice (see Table S2).
To further analyze the differences in metabolized energy, mice of both genetic strains were fasted for two days. Heterozygous animals of both sexes and genetic backgrounds showed a greater loss of body weight than the corresponding controls. This difference was statistically significant for male C3.Dll1tm1Gos/+ and female 129.Dll1tm1Gos/+ mice (Figure 2F and Table S2).
In conclusion, our data suggest that Dll1 heterozygosity causes increased energy requirements.
The cardiovascular mutant phenotype
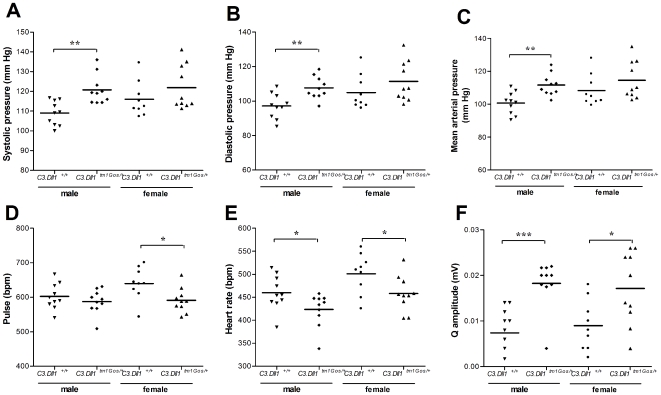
At the age of 14 weeks C3.Dll1tm1Gos/+ and wild-type littermates were analyzed for cardiovascular parameters. The systolic, diastolic and mean arterial pressures were significantly elevated in C3.Dll1tm1Gos/+ male mice (Figure 3A–C). The resting heart rate was determined during blood pressure measurements (“pulse” in beats per minute, bpm) and in anaesthetized mice during electrocardiography (“heart rate” in bpm). In C3.Dll1tm1Gos/+ female animals the average pulse was significantly, in male mice slightly decreased as compared to wild-type littermates (Figure 3D). The heart rate was significantly decreased in both male and female C3.Dll1tm1Gos/+ animals (Figure 3E), indicating a slight bradycardia in heterozygous mutant animals. Moreover, the Q amplitude was significantly increased in the electrocardiogram traces of heterozygotes (Figure 3F).
Figure 3. Selected blood pressure and electrocardiogram parameters in heterozygous C3.Dll1tm1Gos/+ animals and their wild-type littermates.
A–F) Depicted are the scatter dot plot representations and the mean for each group: heterozygous C3.Dll1tm1Gos/+ males (n = 10) and their wild-type littermates (C3.Dll1+/+, n = 10); heterozygous C3.Dll1tm1Gos/+ females (n = 10) and their wild-type littermates (C3.Dll1+/+, n = 9). P-value calculated performing unpaired t-test as variances were not significantly different. P-value: * P<0.05, ** P<0.01, *** P<0.001.
Alteration of clinical chemical blood parameters
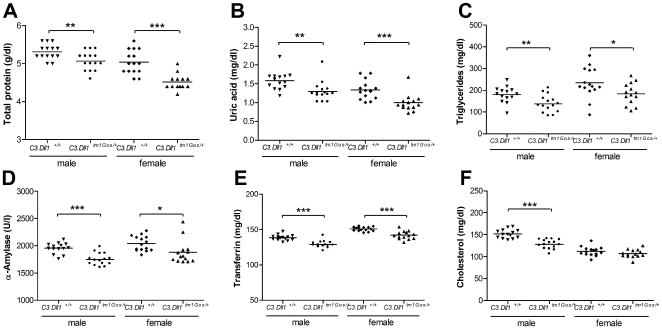
At 12 weeks of age blood samples were taken from fed mice for the analysis of 20 clinical chemical parameters (see Table S3). The concentrations of total protein, uric acid, triglycerides and transferrin, as well as α-amylase activity were significantly decreased in male and female C3.Dll1tm1Gos/+ animals (Figure 4A–E). In addition, several other blood parameters were significantly altered in heterozygous mutant mice of only one gender (see Table S3). For example, the average concentration of cholesterol in the blood of C3.Dll1tm1Gos/+ mice was significantly reduced only in male animals (Figure 4F). Taken together, our data demonstrate that heterozygosity for a Dll1 loss-of-function allele resulted in altered blood concentrations of key metabolites of the protein, carbohydrate and fat metabolism.
Figure 4. Selected clinical chemistry parameters in heterozygous C3.Dll1tm1Gos animals and their wild-type littermates.
A–F) Depicted are the scatter dot plot representations and the mean for each group: heterozygous C3.Dll1tm1Gos/+ males (n = 15) and their wild-type littermates (C3.Dll1+/+, n = 14) and heterozygous C3.Dll1tm1Gos/+ females (n = 14) and their wild-type littermates (C3.Dll1+/+, n = 15). P-value calculated performing unpaired t-test when variances were not significantly different. If significantly different then the Mann-Whitney test was performed. P-value: * P<0.05, ** P<0.01, *** P<0.001.
Immunology screen
The same blood samples as above were analyzed by flow cytometry and indirect ELISA. The population of CD19 positive cells (B-cell population) was significantly increased in female but not male Dll1tm1Gos/+ animals of both genetic backgrounds compared to corresponding wild-type littermates (Table 1 and data not shown). Moreover, the proportion of B1 cells within the B cluster was significantly decreased in female C3.Dll1tm1Gos/+ mice. A reduction of the CD4+ T-cell population in C3.Dll1tm1Gos/+ mice was significant only in females (Table 1). A similar reduction in CD4+ cells was observed in the 129.Dll1tm1Gos/+ genetic background, although the difference between wild-type mice and heterozygous mutants was not statistically significant (data not shown). The population of CD8α,β T-cells showed sex-dependent differences. Compared to the results for the corresponding wild-type littermates this cell population was significantly increased in male and significantly decreased in female heterozygous mutant animals (Table 1). IgG1, IgG2b, IgG3 and IgA parameters were significantly decreased in female C3.Dll1tm1Gos/+ animals. In conclusion, our data suggest that haploinsufficiency of Dll1 causes alterations in immunological parameters, in particular in the number of B-lymphocytes.
Table 1. Immunological parameters in heterozygous C3.Dll1tm1Gos/+ animals.
| Sex | Parameter | C3.Dll1+/+ | C3.Dll1tm1Gos/+ | P-value | Regulation |
| Male | CD19+ (%) | 13.00±1.63 | 12.98±2.60 | 0.98 | ↔ |
| Cd19+/CD5− (%) | 86.33±2.16 | 88.03±1.22 | 0.044* | ↑ | |
| Cd19+/CD5+ (%) | 13.69±2.17 | 11.98±1.21 | 0.043* | ↓ | |
| CD8a+ (%) | 11.67±0.66 | 10.78±1.27 | 0.066 | ↔ | |
| CD4+ (%) | 27.37±3.36 | 25.29±2.34 | 0.13 | ↔ | |
| CD8a/b+ (%) | 9.36±0.92 | 10.67±1.49 | 0.036* | ↑ | |
| Gr-1+ (%) | 8.73±2.17 | 8.96±1.97 | 0.81 | ↔ | |
| γ/δ TCR+ (%) | 0.40±0.20 | 0.48±0.21 | 0.44 | ↔ | |
| IgG1 (µg/ml) | 60.64±30.94 | 82.45±32.25 | 0.085 | ↔ | |
| IgG2a (µg/ml) | 219.08±151.26 | 217.37±147.51 | 0.98 | ↔ | |
| IgG2b (µg/ml) | 78.59±20.35 | 83.78±33.45 | 0.61 | ↔ | |
| IgG3 (µg/ml) | 195.84±88.51 | 180.41±68.46 | 0.60 | ↔ | |
| IgA (µg/ml) | 320.31±190.92 | 333.29±222.32 | 0.87 | ↔ | |
| Anti-DNA (%) | 0.40±0.03 | 0.39±0.04 | 0.56 | ↔ | |
| Rheumatoid. factor (%) | 0.17±0.02 | 0.16±0.02 | 0.51 | ↔ | |
| Female | CD19+ (%) | 13.70±3.57 | 20.28±3.32 | 0.00065*** | ↑ |
| Cd19+/CD5− (%) | 77.84±7.89 | 82.13±2.32 | 0.15 | ↔ | |
| Cd19+/CD5+ (%) | 22.16±7.89 | 17.87±2.32 | 0.15 | ↔ | |
| CD8a+ (%) | 10.26±1.14 | 9.64±1.71 | 0.37 | ↔ | |
| CD4+ (%) | 31.53±4.27 | 22.60±3.49 | 0.00011*** | ↓ | |
| CD8a/b+ (%) | 10.22±1.12 | 8.53±1.09 | 0.0040** | ↓ | |
| Gr-1+ (%) | 8.60±1.96 | 7.86±2.17 | 0.43 | ↔ | |
| γ/δ TCR+ (%) | 0.05±0.02 | 0.05±0.02 | 0.60 | ↔ | |
| IgG1 (µg/ml) | 136.10±53.21 | 72.61±36.40 | 0.00092*** | ↓ | |
| IgG2a (µg/ml) | 331.72±222.39 | 224.28±138.18 | 0.16 | ↔ | |
| IgG2b (µg/ml) | 152.65±69.31 | 94.36±58.59 | 0.022* | ↓ | |
| IgG3 (µg/ml) | 302.33±134.89 | 160.71±80.02 | 0.0021*** | ↓ | |
| IgA (µg/ml) | 377.32±132.22 | 218.62±164.50 | 0.014* | ↓ | |
| Anti-DNA (%) | 0.51±0.08 | 0.44±0.05 | 0.0099** | ↓ | |
| Rheumatoid. factor (%) | 0.22±0.07 | 0.18±0.01 | 0.07 | ↔ |
Values displayed as mean±SD. Data obtained by flow cytometry and indirect ELISA. P-value calculated performing unpaired t-test with samples with equal variances, if not Mann-Whitney test performed: *<0.05, **<0.01, ***<0.001.
Dysmorphology screen
At the age of 9 weeks wild-type and C3.Dll1tm1Gos/+ mice were analyzed for 26 dysmorphology parameters (see Materials and Methods). One wild-type animal out of 58 showed a variant phenotype (kinked tail phenotype), whereas their heterozygous C3.Dll1tm1Gos/+ littermates showed seven variant phenotypes (3 small-body size and 4 kinked tail phenotypes, see Table 2). The Fisher's exact test revealed no significant differences between heterozygous C3.Dll1tm1Gos/+ and wild-type animals in the appearance of a variant morphological phenotype. The reduced body weight and body length of Dll1tm1Gos/+ mice were described above.
Table 2. Skeletal phenotyping of heterozygous Dll1tm1Gos/+ animals.
| Phenotyping screen | Genotype | No variation | Variation | Two-sided P-value |
| Dysmorphology | C3.Dll1+/+ | 58 | 1* | 0.061 |
| C3.Dll1tm1Gos/+ | 52 | 7 | ||
| Quantification lumbar vertebrae | C3.Dll1+/+ | 19 | 0 | 0.678 |
| C3.Dll1tm1Gos/+ | 19 | 1 | ||
| 129.Dll1+/+ | 17 | 2 | ||
| 129.Dll1tm1Gos/+ | 16 | 3 |
Displayed are the numbers of animals showing the observed phenotypes.
One animal was removed of the analysis, as its variant phenotype, an abnormal eye, could be rather due to injury than a genetic component.
The two-sided P-value was calculated using the Fisher's Exact Test or the Chi-square for the genome-wide vs modifier screen, due to the higher number of cases compared.
P-value: *<0.05, **<0.01, ***<0.001.
At 16 to 17 weeks of age 9 to 10 animals of each group (separated by genotype and gender) were investigated with X-ray autoradiography. One heterozygous C3.Dll1tm1Gos/+ animal showed a reduced number of lumbar vertebrae (Table 2). No differences were observed in other parts of the axial skeleton. In the 129X1 background, a different number of lumbar vertebrae was found in heterozygous and wild-type animals. The Fisher's Exact test revealed no significant differences due to the genotype and appearance of an abnormal number of lumbar vertebrae.
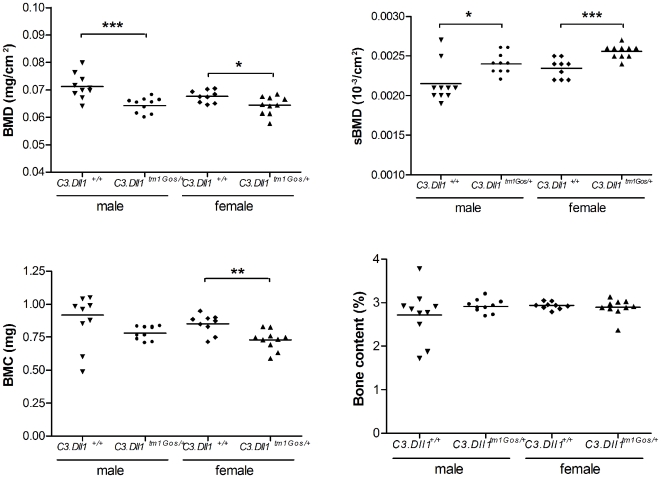
Further bone parameters were measured using DEXA densitometry. Whole body bone mineral density (BMD) was significantly reduced in the C3.Dll1tm1Gos/+ animals compared to their wild-type littermates (Figure 5A). Nevertheless, when related to body weight the specific bone mineral density (sBMD) was significantly increased in C3.Dll1tm1Gos/+ animals (Figure 5B). Whole body bone mineral content (BMC) was significantly reduced in C3.Dll1tm1Gos/+ females, and a similar tendency was observed in male animals (Figure 5C). No differences were found when the bone content was related to body weight (Figure 5D). Comparable results were found for heterozygous mutant and wild-type mice of the 129X1 background (data not shown).
Figure 5. Selected bone parameters obtained with a Dual energy X-ray absorptiometer from heterozygous C3.Dll1tm1Gos/+animals.
A–D) Depicted are the scatter dot plot representations and the mean for each group: heterozygous C3.Dll1tm1Gos/+ males (n = 10) and their wild-type littermates (C3.Dll1+/+, n = 10); heterozygous C3.Dll1tm1Gos/+ females (n = 9) and their wild-type littermates (C3.Dll1+/+, n = 10). P-values calculated performing unpaired t-test when variances were not significantly different. If significantly different then the Mann-Whitney test was performed. P-value: * P<0.05, ** P<0.01, *** P<0.001.
Changes in gene expression patterns
A set of 11 organs from 5 wild-type and 5 heterozygous C3.Dll1tm1Gos/+ male animals was archived for subsequent expression profiling analysis [24]. The brain was selected for the analysis because Dll1 plays an important role in the differentiation of neuronal precursors [25]. Spleen and thymus were chosen based on the requirement of Dll1 signaling for immunological parameters [26], [27]. Finally, gene expression in liver was studied due to the importance of the organ in overall energy metabolism. In brain and spleen no significantly regulated genes were detected. 13 significantly downregulated genes were found in the liver of C3.Dll1tm1Gos/+ animals (Table 3). No up-regulated genes were identified in this organ. Among the regulated genes several are associated with metabolic liver functions, like cholesterol biosynthesis.
Table 3. Genes regulated in heterozygous C3.Dll1tm1Gos/+ animals when compared to wild-type littermates.
| Gene symbol | Entrez ID | Name | Function | Mean log2 ratio |
| Genes regulated in the liver | ||||
| Cdk2 | 12566 | Cyclin-dependent kinase 2 | Regulation of cell cycle | −2.24 |
| Cth | 107869 | Cystathionase | Methionine metabolism | −2.10 |
| Idi1 | 319554 | Isopentenyl-diphosphate delta isomerase | Cholestrol biosynthesis | −1.93 |
| Serpina3k | 20714 | Serine peptidase inhibitor, clade A, member 3K | Peptidase inhibitor | −1.89 |
| Cul2 | 71745 | Cullin 2 | E3 ubiquitin ligase complex | −1.88 |
| Serpina3c | 16625 | Serine peptidase inhibitor, clade A, member 3C | Peptidase inhibitor | −1.87 |
| Bhmt | 12116 | Betaine-homocysteine methyltransferase | Methionine metabolism | −1.87 |
| Armet | 74840 | Arginine-rich, mutated in early stage tumors | Unknown | −1.82 |
| Serpina3h | 546546 | Serine peptidase inhibitor, clade A, member 3H | Peptidase inhibitor | −1.84 |
| Siah1a | 20437 | Seven in absentia 1A | E3 ubiquitin ligase complex | −1.84 |
| Cyp2f2 | 13107 | Cytochrome P450, family 2, subfamily f, polypeptide 2 | Metabolism of xenobiotics | −1.82 |
| Hmgcs1 | 208715 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | Cholesterol biosynthesis | −1.71 |
| Cyp2c70 | 226105 | Cytochrome P450, family 2c, polypeptide 70 | Metabolism of xenobiotics | −1.70 |
| Genes regulated in the thymus | ||||
| mt-Rnr2 | 16S rRNA, mitochondrial | Mitochrondrial RNA | −2.68 | |
| 2310043N10Rik | 66961 | Unknown | −2.20 | |
| Tcrb-V13 | 269846 | T-cell receptor beta, variable 13 | T cell development | −2.11 |
| Bcl11b | 58208 | B-cell leukemia/lymphoma 11B | T cell development | −2.91 |
| Vldlr | 22359 | Very low density lipoprotein receptor | Cholesterol metabolism | −2.90 |
| Sfrs5 | Splicing factor, arginine/serine-rich 5 | RNA metabolic process | −1.69 | |
| Ddx6 | 13209 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | RNA metabolic process | −1.63 |
| Higd1b | 75689 | HIG1 domain family, member 1B | Unknown | −1.67 |
| AK160221 | AK160221 | Unknown | −1.62 | |
| Apc | 11789 | Adenomatosis polyposis coli | T-cell development | −1.61 |
| Trip12 | 14897 | Thyroid hormone receptor interactor 12 | E3 ubiquitin ligase complex | −1.52 |
| EU433307 | Unknown | −1.50 | ||
| Akap8 | 56399 | A kinase (PRKA) anchor protein 8 | Unknown | −1.49 |
| Ywhab | 54401 | Tyrosine 3-monooxygenase protein, beta polypeptide | T-cell development | −1.49 |
In the thymus of heterozygous mutant animals no up-regulated genes were detected. Instead, 15 genes were significantly downregulated in heterozygous C3.Dll1tm1Gos1/+ animals (Table 3). One third are associated with T cell differentiation or involved in the progress of maturation or migration of T cells and several have unknown functions. Some of the genes regulated in the thymus of heterozygous mutant animals were also found to be regulated in homozygous mutant embryos such as Ddx6, Higd1b and polypeptides of Ywha (14-3-3) complex ([28] and unpublished data).
Variant phenotype of the missense mutant line C3.Dll1C413Y/+
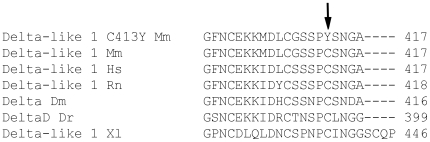
The C3.Dll1C413Y/+ mutant line was isolated from an ENU mutagenized combined sperm and genome archive by sequencing the Dll1 coding region [29]. C3.Dll1C413Y/+ mutants have a missense mutation in exon 8 which leads to the exchange of the cysteine in position 413 by a tyrosine. This mutation lies within the predicted 6th EGF-like repeat, which is highly conserved from insects to vertebrates (Figure 6).
Figure 6. Partial protein sequence alignment of Delta and Delta-like-1 proteins.
The sequence alignment was performed with ClutsalW2 [90]. The missense mutation C413Y is indicated by an arrow. The species used for these alignment are: Delta-like 1 Mm, Delta-like 1 Mus musculus NP_031891.2; Delta-like 1 Hs, Delta-like 1 Homo sapiens NP_114452.1; Delta-like 1 Rn, Delta-like 1 Rattus norvegicus NP_005609.3; Delta Dm, Delta Drosophila melanogaster NP_477264.1; DeltaD Dr, DeltaD Danio rerio; Delta-like 1 Xl, similar to Delta-like 1 Xenopus laevis.
Unlike homozygous Dll1tm1Gos mutants, homozygous C3.Dll1C413Y/C413Y mutants are viable, although only 32% of the expected homozygous animals are alive after weaning. Dll1C413Y/C413Y animals are lighter than their wild-type and heterozygous littermates and are generally in a bad health condition. Most of them died or had to be sacrificed within the first months of life. Since we suspect that the Dll1C413Y mutation is a partial loss-of-function, we compared the phenotype of Dll1C413Y/+ with Dll1tm1Gos/+ animals (Table 4). Several phenotypes of Dll1tm1Gos/+ mice were also found in Dll1C413Y/+ mice. For example, the body weight and length of heterozygous C3.Dll1C413Y/+ animals was decreased comparable to C3.Dll1tm1Gos/+ mice. In addition, the C3.Dll1C413Y/+ animals show a significantly reduced bone mass density and bone mineral content. The fat mass was also significantly decreased in C3.Dll1C413Y/+ animals. Also comparable to the changes observed in Dll1tm1Gos/+ animals, the systolic, diastolic and mean arterial pressures were significantly increased, and the pulse significantly decreased in C3.Dll1C413Y/+ animals. However, lean mass, locomotor activity, and metabolized energy were increased in Dll1C413Y/+ compared to wild-type littermates. This is somewhat in contrast to Dll1tm1Gos/+ animals, which showed a significant decrease in lean mass and a tendency towards reduced locomotor activity. Concerning the clinical chemical parameters both mutant lines showed similar tendencies in the concentrations of cholesterol and triglycerides in their blood plasma. Whereas Dll1tm1Gos/+ animals showed decreased concentrations of urea, this concentration was significantly increased in the blood of Dll1C413Y/+ animals. Finally, in contrast to the Dll1tm1Gos/+, no differences were observed in populations of CD19+ and CD4+ cell lines between the C3.Dll1C413Y/+ and wild-type littermates.
Table 4. Comparison of the variant phenotypes due to the C3.Dll1tm1Gos and C3.Dll1C413Y alleles.
| Phenotyping screen | Parameters | Male C3.Dll1tm1Gos mutant/wt (%) | Male C3.Dll1C413Y mutant/wt (%) | Female C3.Dll1tm1Gos mutant/wt (%) | Female C3.Dll1C413Y mutant/wt (%) |
| Dysmorpho-logy | Body weight (g) | −19.9*** | −9.0** | −12.9*** | −14.9*** |
| Body length (cm) | −4.0* | −2.4* | −3.9*** | −4.3*** | |
| DEXA densitometry | Bone mineral density (mg/cm2) | −9.9*** | −9.7** | −4.5* | −8.2** |
| Bone mineral content (mg) | −14.9n.s. | −12.9n.s. | −14.0** | −25.3* | |
| Fat mass (g) | −37.1* | −31.8* | −15.3n.s. | −52.2** | |
| Lean mass (g) | −12.8* | 5.8n.s. | −12.7*** | 29.8* | |
| Neurology | Locomotor acitivity (squares) | −14.9n.s. | 45.8n.s. | −10.2n.s. | 75.0* |
| Metabolism | Food consumption (g/day) | −7.0** | 2.2n.s. | −22.5** | 0n.s. |
| Metabolized energy (kJ/g×day) | 22.8*** | 12.9*** | 6.6n.s. | 17.8*** | |
| Cardiovascular | Systolic pressure (mmHg) | 10.8** | 6.0n.s. | 5.2n.s. | 10.9* |
| Diastolic pressure (mmHg) | 10.8* | 7.2n.s. | 6.0n.s. | 12.4* | |
| Mean arterial pressure (mmHg) | 10.8* | 6.8n.s. | 5.7n.s. | 12.0* | |
| Pulse (bpm) | −2.4n.s. | −2.0n.s. | −7.6* | −1.2n.s. | |
| Clinical chemistry | Urea (mg/dl) | −11.2** | 9.1** | −1.5n.s. | 15.3*** |
| Cholesterol (mg/dl) | −16.2*** | −8.8* | −4.6 | −4.7n.s. | |
| Triglyceride (mg/dl) | −23.4** | −35* | −21.1* | −4.4n.s. |
%: Difference of the means between mutant animals and wild-type in percent. *,**,*** indicates when in the statistical analysis the means were found to be significantly different with a probability of *<0.05. **<0.01 and ***<0.001, n.s. equals means not statistically significantly different.
Discussion
The comprehensive investigation of Dll1 haploinsufficiency reveals a complex phenotype, indicating that several biological processes are affected.
Immunological phenotype
One of the most studied roles of Notch signaling in adult stages is its involvement in T/B-cell commitment (for review see [26], [27]). Despite a high number of studies, all mechanisms in this process still have not been elucidated. In particular, the role of the Notch ligands is less understood.
Hematopoietic stem cells of the bone marrow give rise to CLP, further proB cells, and immature B cells (in the case that Notch1 is inactive). These cells migrate to the spleen where differentiation occurs. Although we did not detect any differentially regulated gene in the spleen, the frequencies of B- and T-cell populations in the blood of heterozygous animals show a sex-dependent phenotype. As in wild-type mice of different strains under baseline conditions the frequency of T cells is generally higher in females [30], we could interpret our findings as a loss of sex-difference in the size of the T-cell compartment. Whether this might hind to a link of Dll1 to sex-dependent regulatory mechanisms remains unclear.
Hozumi et al. [31] previously demonstrated that conditional ablation of Dll1 in hematopoietic cells leads to a disappearance of the marginal zone in the spleen. In the spleen, type 1 transitional B-cells (B1) differentiate into follicular and marginal zone cells. Although not directly investigated, our data affirm the role of Delta-like 1 in B cell commitment and maturation.
The CD8αβ+ T cell blood profile of the mutant Dll1 looks ambiguous. CD8αβ+ T cells are significantly increased in male animals but significantly decreased in heterozygous females. Consistent with findings for Notch signaling in lymphocyte development [26], [27] γδ-TCR+ cell numbers were not changed in homozygous mutants. Epigenetics has been identified as mechanism in T-cell lineage commitment [32], [33], and could therefore explain the ambiguous variant phenotype observed here.
Nevertheless, Dll1 haploinsufficiency leads to alterations in the expression of genes, which are clearly linked to T cell differentiation and maturation. For example, Apc and Bcl11b are downregulated in the thymus. Both genes are essential in maintaining the DN3 checkpoint status, when the cell fate declines either to be αβ- or γδ-T cells [34]–[36]. In the DN3 stage, TCRβ allelic exclusion plays a pivotal role, which is initiated by the pre-TCR complex and controlled by Vβ gene rearrangements. Another gene of the Vβ cluster, Tcrb-V13, which is required for efficient Vβ gene rearrangements [37], was also downregulated. Additionally, two further genes, CD8α and Ywhab, which are associated with T cell differentiation processes [38], [39], were found to be downregulated in Dll1 haploinsufficient animals.
The blood profile showed reduced CD4+ T-cell levels in heterozygous animals, which are significantly decreased only in females. Although a role for Notch signaling in CD8+-T versus CD4+-T commitment is already described [27], the specific role for Dll1 remains unclear. We found a downregulation of Trip12 gene expression in thymocytes. Trip12 is a hect-protein with possibly E3 ubiquitin ligase function [40]–[42]. It is already known, that Notch signaling is highly regulated by ubiquitinylation (for review see [43]). Recently it was shown that double mutants of Notch and Itch, another E3 ubiquitin ligase from the hect-family, display severely perturbed thymocyte development [44], [45]. The kind of interaction between Trip12 and Dll1 has to be elucidated.
In summary, the blood profile and the thymus transcriptomic data affirm an essential role for Dll1 in lymphoid cell differentiation, maturation and function. Dll1 haploinsufficiency of the whole organism induces rather a subtle phenotype and the functionality of the Dll1C413Y allele seems to be sufficient to maintain a normal network function. Probably Dll1 haploinsufficiency has no immunological pathological consequences, unless the animals are subjected to an immunological challenge. This is in accordance with recent publication, where administration of Delta-Fc (Delta-like 1-immunoglobulin Fc fusion protein) to the lung had an impact on the inhibition of immunological responsiveness, but only in sensitized and challenge animals [46].
Metabolic phenotype
Dll1 haploinsufficiency leads to lighter and smaller mice with altered fat and lean mass ratio, higher energy uptake and metabolized energy when normalized to body weight. Hyperactivity seems not to be the reason for the higher energy expenditure. The hyperactive phenotype of C3.Dll1C413Y/+ and 129.Dll1tm1Gos/+ females is probably related to a neurological phenotype that should be investigated in detail elsewhere.
Increased blood pressure in haploinsufficient animals may be explained by increased energy demands. Although we did not determine the basal metabolic rate, clinical-chemical parameters in plasma and liver transcriptomic analysis indicate that Dll1 haploinsufficiency causes alterations of the entire metabolism. The downregulation of genes in the liver is most likely caused by external signals since Dll1 itself is not expressed in this organ [31], [47]. Thus, the downregulated genes found must be a consequence of signals coming from outside the liver.
Total protein levels in blood plasma were significantly reduced in Dll1 mutants. For example, the plasma concentration of transferrin, one of the main plasma proteins, was decreased due to Dll1 haploinsufficiency. Interestingly, nearly all examined immunoglobulins were significantly reduced only in female mutants. The reduction of total protein levels in plasma is subtle and mainly due to the reduction of selected proteins and not caused by a liver disease since the levels of the enzymes alanine-aminotransferase and aspartate-aminotransferase were not altered.
The transcriptional levels of cystathionase (Cth) and betaine-homocysteine methlytransferase (Bhmt), two enzymes involved in amino acid metabolism, were altered. Cth catalyzes one step in the reactions from homocysteine to cysteine; Bhmt creates methionine from homocysteine. Alterations in the function of both enzymes have been described to alter homocysteine levels [48]–[50]. We hypothesize that a reduction in these transcript levels leads to a reduction of homocysteine levels in plasma rather than to an altered protein metabolism. Consistent with reports on the influence of altered homocysteine levels on bone stability [51], [52] we found a reduced bone mineral density in Dll1 haploinsufficient animals. Recently it was reported that homocysteine is able to modulate Notch1 function by interacting with cystein bonds in the Notch receptor [50]. Since the Delta-like 1 does not possess cysteine-rich regions the link between the Notch ligand and homocysteine might be indirect.
Dll1 haploinsufficiency alters also the lipid metabolism. For example, triglycerides and cholesterol are decreased in plasma of Dll1tm1Gos/+ animals. Reduced triglycerides may indicate subtle alterations in lipid handling. Cyp2c70, downregulated in the liver of Dll1 mutants, is involved in linolenic and arachidonic acid metabolism. Of more interest is the downregulation of Idi1 and Hmgcs1, two genes which encode for enzymes involved in cholesterol biosynthesis [53], [54]. Interestingly, low levels of cholesterol leads to high expression of Adam10, coding for a protease also known as Kuzbanian, which is essential for Notch activation [55]. Moreover, when Adam10 is not expressed Dll1 is overexpressed [56]. Recently, it has been shown that cholesterol is a modulator of γ-secretase activity, and therefore modulates Notch signaling as well [57]. Our results suggest a reciprocal regulation. A decrease in Dll1 gene expression leads to low cholesterol levels via transcriptional regulation.
Previously, we presented six lines out of our modifier screen with either high cholesterol or transferrin or total protein levels or with altered T- and B-cell patterns, that were all linked to Dll1 haploinsufficiency [14]. Further studies on these lines will lead to the discovery of several Notch-signaling suppressors.
Skeletal phenotype
Dll1 haploinsufficiency leads to a reduction in femoral and whole-body bone mineral density as well as bone mineral content. The role of Notch-signaling pathway components in bone remodeling is known [58]. A close relationship between body weight and bone mass was reported for humans. It was also shown that bone mineral density correlates with metabolic rate and that there is a close connection between lean mass and bone mineral density [59]–[64]. Recent studies in mice demonstrate a reciprocal hormonal link between energy metabolism and bone remodeling [65], [66]. In conclusion, the reduced bone mineral density and bone mineral content in Dll1 mutants is most probably caused by a reduced body size and an altered metabolic rate.
Cordes et al. reported about homeotic transformations with incomplete penetrance in the cervical region of heterozygous Dll1 mutants [67]. Here we did not observe any alterations in the number of vertebrae linked to Dll1 haploinsufficiency, due to the small number of animals studied.
EGF-repeat dependent phenotype
The exchange of an amino acid in the 6th EGF-repeat of the Delta-like 1 protein (C413Y) leads to a phenotype that is, except for alterations in the B- and T-cell patterns, almost identical to the complete loss of an entire Dll1 allele. The fact that at least a few homozygous C3.Dll1C413Y/C413Y mutants were born suggests variable doses of functional Delta-like 1 protein.
Proteolysis mediated Notch signaling regulation
The Notch signaling pathway is mediated by proteolysis and does not seem to be mediated by second messengers (for review see [2]). But some questions remain open. So it is still unclear which E3 ubiquitin ligase monoubiquitinates Notch prior to γ-secretase cleavage [3]. We have already mentioned that Dll1 haploinsufficiency leads in the thymus to downregulation of a protein that shows E3 ubiquitin ligase function, Trip12. However, this is not the only gene found to be downregulated that belongs to the ubiquitin machinery. For example, Siah1, which belongs to the E3 ubiquitin ligase complex, is known to promote degradation of Numb, and thereby leads to activation of Notch [68]–[71]. Cul2, another protein of the E3 ubiquitin ligase complex, degrades HIF-1α, whose interaction with NICD leads to transcriptional activation of Notch targets [72]–[74]. As Notch signaling is fine-tuned by different proteases, it can be regulated also by proteases inhibitors. Here we found that due to Dll1 haploinsufficiency three of the 14 components of the human SERPINA3 (α1-antichymotrypsin) homolog cluster were downregulated, Serpina3c, Serpina3h and Serpina3k. Other components of the cluster were not found to be differentially regulated, like Serpina3a and Serpina3g. Serpina3h and Serpina3k probably encode protease inhibitors, whereas Serpina3c is more likely an ortholog of kallistatin in humans [75]. Kallistatin is known to have an influence on blood pressure [76], [77].
Taken together, Dll1 haploinsufficiency leads to a downregulation of selected genes that could function as modulators of the Notch signaling pathway, probably due to feedback mechanisms in maintaining the fine-tuning of this signaling network.
Link to Wnt signaling
Interactions between Notch and Wnt signaling were first uncovered in the developing wing disc of the fly (for review see [78]). In vertebrates, mutually dependent interactions between Notch and Wnt signaling have been described in several processes like somitogenesis or T-cell development [26], [78]. Wnt signaling activates usually the expression of Notch ligands. The recurrence of this regulatory relationship is higher than the occurrence of regulation in two distinct interacting networks, suggesting that probably Wnt and Notch signaling devices are part of an integrative system called ‘Wntch’ [78]. It is not the aim of this study to investigate this hypothesis. Nevertheless, we detected in this work a relation between the Notch signaling and the Wnt signaling pathway. The two genes, Apc and Siah1, which were found to be downregulated in Dll1 mutants, are known to act via β-catenin and by extension on Wnt signaling [34], [35], [70].
Conclusion
Dll1 haploinsufficiency leads to a complex phenotype with several biological processes altered. These alterations emphasize the importance of Dll1 mainly in metabolism, energy balance and in immunology. The Dll1 haploinsufficient animals are smaller, lighter, with altered fat to lean ratio and have an increased blood pressure and a slight bradycardia. At the immunological level a subtle phenotype is observed due to the effects and fine-tuning of the signaling network at different levels of differentiation, proliferation and function of lymphocytes. Moreover, the importance of the proteolytic regulation of the Notch signaling network is emphasized.
Materials and Methods
Mice
Mice were housed and handled according to the federal animal welfare guidelines and all the animal studies were approved by the state ethics committee. Mouse husbandry was conducted under a continuously controlled specific pathogen-free (SPF) hygiene standard according to the Federation of European Laboratory Animal Science Associations (FELASA). For this study we used mice carrying the Dll1tm1Gos allele on a 129X1/SvJ background [15], and these mice were outcrossed 11 generations to wild-type C3HeB/FeJ mice. The mutant line was further maintained by mating between siblings. The C3.Dll1C413Y/+ mutant line was isolated from an ENU mutagenized combined sperm and genome archive by sequencing the Dll1 coding region [29]. Phenotypic screening was performed by comparing heterozygous mice with the same number of age- and sex-matched littermate controls [20]. Mice were genotyped for the Dll1-LacZ insertion as described elsewhere [14]. The isogenicity of the Dll1tm1Gos in the C3HeB/FeJ background was evaluated with the SNP panel developed at our institute for linkage analysis as described elsewhere [14].
Metabolic screen
At the age of 17 to 19-weeks wild-type and heterozygous Dll1tm1Gos mutants entered the metabolic screen in the GMC, and were analyzed as described previously [79]–[81]. In the primary screen mice were single-caged and fed ad libitum for a period of 14 days. The following parameters were measured: body weight, food consumption (Fcon), rectal temperature (Tre), daily faeces production (Fec), energy uptake (Eup), energy content of the faeces (Efec), metabolizable energy (Emet) and food assimilation coefficient (Fass). In the tertiary screen the body composition was analyzed. At a mean of 31 weeks of age the animals were killed, weighed and the gastrointestinal tract was removed. Body composition was determined by drying the dissected carcass, weighing the dry carcasses and fat mass was determined by extracting the lipids using a refluxing Soxhlet apparatus with petrol ether as solvent for 16 hours [82]. The post extraction dry mass was determined. Lean mass was calculated as the carcass weight minus the fat mass.
Cardiovascular screen
At the age of 14 weeks the mice entered the cardiovascular screen of the GMC [83]. Blood pressure was measured in conscious mice with a non-invasive tail-cuff method using the MC4000 Blood pressure Analysis Systems (Hateras Instruments Inc.). Pulse detection, cuff inflation and pressure evaluation were automated by the system software. At least 20 to 48 individual measurements were pooled to obtain a mean over the four measurements days for each animal. The electrocardiogram analysis (ECG) was performed in isoflurane-anesthetized mice by the use of three metal bracelets on the joints of the feet. Positioning on the front paws and the left hind-paw allows the recording of the bipolar standard limbs I, II, and III and the augmented unipolar leads AVF, AVR and AVL. The ECG was recorded for seven minutes. In the quantitative ECG analysis sets of 5 analyzed beats were averaged for each animal. The shape analysis of the ECG traces was performed with the software ECG-auto (EMKA technologies).
Clinical chemistry screen
Blood samples were obtained from 12-week-old mice by puncturing the retro-orbital sinus under anesthesia [84]. Plasma was separated by centrifugation (10 min, 4656×g; Biofuge, Heraeus; Hanau, Germany) diluted 1∶2 with deionized water and analyzed in the GMC for clinical chemistry parameters. The 20 parameters measured cover a broad spectrum suitable to investigate alterations in metabolism, organ function and electrolyte homeostasis. In detail the electrolytes sodium, potassium, total calcium, chloride and inorganic phosphorus; total protein in plasma and the proteins ferritin and transferrin; the metabolites creatinine, urea, uric acid, cholesterol, triglycerides and glucose; and the enzymes creatine kinase, alanine-aminotransferase (ALT), aspartate-aminotransferase (AST), alkaline phosphatase, α-Amylase and lipase were analyzed. The parameters were analyzed using an Olympus AU 400 autoanalyzer and adapted reagents from Olympus (Hamburg, Germany), as described elsewhere [85]. The results presented here include individuals investigated under the same analytical conditions.
Immunology screen
A fraction of the blood sample taken at 12 weeks of age was separated to analyze immunological parameters using ELISA and flow cytometry, as described previously [86]. The following main cell populations were analyzed by flow cytometry: B cells (CD19+ clone 1D3), B1 B cells (CD19+CD5+, clone 53-7.3), B2 B cells (CD19+CD5−), T cells (CD3+, clone 145-2C11), CD4+ T cells (clone RM4-5), CD8+ T cells (CD8α, clone 53-6.7; CD8β, clone H35-17.2), γ/δT cells (clone GL3), granulocytes (Gr-1+, clone RB6-8C5), and NK cells (CD49b+, clone DX5). Data were acquired on a LSR II flow cytometer (Becton Dickinson, USA) and were analyzed using FlowJo software (TreeStar Inc, USA). All samples were acquired until a total number of 30,000 cells were reached.
The plasma levels of IgM, IgG1, IgG2a, IgG2b, IgG3, and IgA were determined simultaneously in the same sample using a bead-based assay with monoclonal anti-mouse antibodies conjugated to beads of different color regions (Biorad, USA), and acquired on a Bioplex reader (Biorad). The presence of rheumatoid factor and anti-DNA antibodies was evaluated by indirect ELISA with rabbit IgG (Sigma-Aldrich, Steinheim, Germany) and calf thymus DNA (Sigma-Aldrich), respectively, as antigens and AP-conjugated goat anti-mouse secondary antibody (Sigma-Aldrich). Serum samples from MRL/MpJ-Tnfrsf6lpr mice (Jackson Laboratory, Bar Harbor, USA) were used as positive controls in the autoantibody assays.
Dysmorphology screen
The animals were checked for general condition and health at the age of 5 weeks when entering the GMC. At the age of nine weeks mice were subjected to morphological observation following the protocol described previously [87]. Growth, weight, body size, eye, coat hair growth, coat hair texture, hair follicle structure and orientation, skin pigmentation, skin texture and condition of vibrissae, limbs, digits, tail, teeth, ear morphology, musculature, seizures and epilepsy, motor capabilities and coordination, movement, feeding and drinking behavior, respiratory system, reproductive system or other abnormalities in the body morphology were investigated. X-ray analysis, as well as DEXA scans were performed at the age of 16 weeks. For the X-ray analysis animals were analyzed with the Faxitron X-ray Model MX-20 (Specimen Radiography System, USA). To check bone density alterations the mice were investigated with the pDEXA Sabre X-ray Bone Densitometer (Norland Medical Systems. Inc., UK).
Molecular phenotyping
At the age of 16 weeks five wild-type and five heterozygous Dll1tm1Gos/+ mutants were killed between 9 and 12 am by carbon dioxide and gene expression in liver, spleen, thymus and brain was analyzed as described previously [24]. Briefly, RNA was extracted using the RNeasy Midi kit (Quiagen) following the manufacturer's instructions. For each cDNA array, 15 µg of total RNA were used for reverse transcription and indirect labeled with the fluorescent dyes. Cy3 or Cy5 (Amersham Biosciences, Freiburg, Germany), according to a modified TIGR protocol [88]. In total our cDNA chips contains 20355 spotted sequences (for a full description see GEO database GPL3697). All experiments were submitted to the GEO database: GSE11867. After hybridization the dried slides were scanned with a GenePix 4000A microarray scanner, and the images were analyzed using the GenePix Pro 6.0 image processing software (Axon Instruments). Statistical analyses were performed using TM4 microarray suite [24], [89].
Data analysis
For biological interpretation of the data the software tools MetaCore™ (GeneGo Inc., St. Joseph, MI, USA) and BiblioSphere PathwayEdition™ Genomatix Software GmbH, Munich, Germany) were used. The databases Mouse Genome Informatics (MGI, The Jackson Laboratory, USA), MapViewer (NCBI, USA), GeneCards® (Weizmann Institute of Science, Israel), The Kyoto Encyclopedia of Genes and Genomes (KEGG, Kanehisa Laboratories, Japan) and The Comprehensive Enzyme Information System (BRENDA, TU Braunschweig, Germany) were used to further interpret the biological meaning of the data.
Statistical analyses
All statistical tests were performed using either the software package GraphPad Prism 4.01 (GraphPad Software, USA) or the R Version 2.7.0 for Windows provided by the R Foundation of Statistical Computing. Outliers defined as those falling outside the 25th or 75th quantile and 1.5 times the interquantile range were discarded. The distribution of the data was assessed using the Shapiro-Wilk normality test. The variances were compared using the Levene Test. Samples with continuous data were compared using the two-sample unpaired t-test or the Mann-Whitney test. Samples with categorical data where compared using the Fisher's Exact Test or the Chi-square Test.
Supporting Information
(0.05 MB DOC)
(0.04 MB DOC)
(0.06 MB DOC)
Acknowledgments
We kindly thank the members of the German Mouse Clinic for comprehensive phenotyping of the mutant mouse lines and fruitful discussions and Dian Soewarto for scientific discussion. We kindly thank Tomek Mijalski for the outcrossing of the Dll1tm1Gos mouse line to C3HeB/FeJ background and Matthias Klaften for investigating the isogenicity of the C3.Dll1tm1Gos/+ mouse line. We kindly thank Susanne Axtner, Miriam Backs, Gerlinde Bergter, Silvia Crowley, Nicole Ehrhardt, Sandra Hoffmann, Elfi Holupirek, Kerstin Kutzner, Andreas Mayer, Marcel Schieven, Sandra Schädler, Reinhard Seeliger, Magdalena Trochimiuk and Susanne Wittich for technical assistance. We also kindly thank our animal caretaker team. Data will be published in www.europhenome.org.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was further supported by NGFN plus grants from the Bundesministerium für Bildung und Forschung (01GS0850 (WH, MH, SH, WW, HF, VG-D, and MHA), 01GS0851 (L.B., BR, EW and Th.K.), 01GS0869 (J.R., M.K.), 01GS0854 (A.S. and B.I.), 01GS0852 (TA and DB) and by an EU grant (LSHG-2006-037188, German Mouse Clinic). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kadesch T. Notch signaling: the demise of elegant simplicity. Curr Opin Genet Dev. 2004;14:506–512. doi: 10.1016/j.gde.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Ilagan MX, Kopan R. SnapShot: notch signaling pathway. Cell. 2007;128:1246. doi: 10.1016/j.cell.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Nichols JT, Miyamoto A, Weinmaster G. Notch signaling–constantly on the move. Traffic. 2007;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- 4.Harper JA, Yuan JS, Tan JB, Visan I, Guidos CJ. Notch signaling in development and disease. Clin Genet. 2003;64:461–472. doi: 10.1046/j.1399-0004.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- 5.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 6.Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134:2709–2718. doi: 10.1242/dev.004184. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann JJ, Iruela-Arispe ML. Notch signaling in blood vessels: who is talking to whom about what? Circ Res. 2007;100:1556–1568. doi: 10.1161/01.RES.0000266408.42939.e4. [DOI] [PubMed] [Google Scholar]
- 8.Kohler C, Bell AW, Bowen WC, Monga SP, Fleig W, et al. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 2004;39:1056–1065. doi: 10.1002/hep.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leker RR. Manipulation of endogenous neural stem cells following ischemic brain injury. Pathophysiol Haemost Thromb. 2006;35:58–62. doi: 10.1159/000093545. [DOI] [PubMed] [Google Scholar]
- 10.Morrissey J, Guo G, Moridaira K, Fitzgerald M, McCracken R, et al. Transforming growth factor-beta induces renal epithelial jagged-1 expression in fibrotic disease. J Am Soc Nephrol. 2002;13:1499–1508. doi: 10.1097/01.asn.0000017905.77985.4a. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T, Terada Y, Kuwana H, Tanaka H, Okado T, et al. Expression and function of the Delta-1/Notch-2/Hes-1 pathway during experimental acute kidney injury. Kidney Int. 2008;73:1240–1250. doi: 10.1038/ki.2008.74. [DOI] [PubMed] [Google Scholar]
- 12.Siveke JT, Lubeseder-Martellato C, Lee M, Mazur PK, Nakhai H, et al. Notch signaling is required for exocrine regeneration after acute pancreatitis. Gastroenterology. 2008;134:544–555. doi: 10.1053/j.gastro.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, et al. Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci U S A. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubio-Aliaga I, Soewarto D, Wagner S, Klaften M, Fuchs H, et al. A genetic screen for modifiers of the delta1-dependent notch signaling function in the mouse. Genetics. 2007;175:1451–1463. doi: 10.1534/genetics.106.067298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrabe de Angelis M, McIntyre J, 2nd, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- 16.Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 17.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 18.Lardelli M, Williams R, Mitsiadis T, Lendahl U. Expression of the Notch 3 intracellular domain in mouse central nervous system progenitor cells is lethal and leads to disturbed neural tube development. Mech Dev. 1996;59:177–190. doi: 10.1016/0925-4773(96)00589-8. [DOI] [PubMed] [Google Scholar]
- 19.Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, et al. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gailus-Durner V, Fuchs H, Becker L, Bolle I, Brielmeier M, et al. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat Methods. 2005;2:403–404. doi: 10.1038/nmeth0605-403. [DOI] [PubMed] [Google Scholar]
- 21.Brown SD, Chambon P, Hrabé de Angelis M. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet. 2005;37:1155. doi: 10.1038/ng1105-1155. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs H, Lisse T, Hans W, Abe K, Thiele F, et al. Phenotypic characterization of mouse models for bone-related diseases in the German Mouse Clinic. J Musculoskelet Neuronal Interact. 2008;8:13–14. [Google Scholar]
- 23.Kiernan AE, Li R, Hawes NL, Churchill GA, Gridley T. Genetic background modifies inner ear and eye phenotypes of jag1 heterozygous mice. Genetics. 2007;177:307–311. doi: 10.1534/genetics.107.075960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horsch M, Schadler S, Gailus-Durner V, Fuchs H, Meyer H, et al. Systematic gene expression profiling of mouse model series reveals coexpressed genes. Proteomics. 2008;8:1248–1256. doi: 10.1002/pmic.200700725. [DOI] [PubMed] [Google Scholar]
- 25.Grandbarbe L, Bouissac J, Rand M, Hrabe de Angelis M, Artavanis-Tsakonas S, et al. Delta-Notch signaling controls the generation of neurons/glia from neural stem cells in a stepwise process. Development. 2003;130:1391–1402. doi: 10.1242/dev.00374. [DOI] [PubMed] [Google Scholar]
- 26.Ciofani M, Zuniga-Pflucker JC. The thymus as an inductive site for T lymphopoiesis. Annu Rev Cell Dev Biol. 2007;23:463–493. doi: 10.1146/annurev.cellbio.23.090506.123547. [DOI] [PubMed] [Google Scholar]
- 27.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 28.Machka C, Kersten M, Zobawa M, Harder A, Horsch M, et al. Identification of Dll1 (Delta1) target genes during mouse embryogenesis using differential expression profiling. Gene Expr Patterns. 2005;6:94–101. doi: 10.1016/j.modgep.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Augustin M, Sedlmeier R, Peters T, Huffstadt U, Kochmann E, et al. Efficient and fast targeted production of murine models based on ENU mutagenesis. Mamm Genome. 2005;16:405–413. doi: 10.1007/s00335-004-3028-2. [DOI] [PubMed] [Google Scholar]
- 30.Petkova SB, Yuan R, Tsaih SW, Schott W, Roopenian DC, et al. Genetic influence on immune phenotype revealed strain-specific variations in peripheral blood lineages. Physiol Genomics. 2008;34:304–314. doi: 10.1152/physiolgenomics.00185.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, et al. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 32.Wilson CB, Makar KW, Shnyreva M, Fitzpatrick DR. DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin Immunol. 2005;17:105–119. doi: 10.1016/j.smim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 34.Gounari F, Chang R, Cowan J, Guo Z, Dose M, et al. Loss of adenomatous polyposis coli gene function disrupts thymic development. Nat Immunol. 2005;6:800–809. doi: 10.1038/ni1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukushima T, Zapata JM, Singha NC, Thomas M, Kress CL, et al. Critical function for SIP, a ubiquitin E3 ligase component of the beta-catenin degradation pathway, for thymocyte development and G1 checkpoint. Immunity. 2006;24:29–39. doi: 10.1016/j.immuni.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 37.Ryu CJ, Haines BB, Lee HR, Kang YH, Draganov DD, et al. The T-cell receptor beta variable gene promoter is required for efficient V beta rearrangement but not allelic exclusion. Mol Cell Biol. 2004;24:7015–7023. doi: 10.1128/MCB.24.16.7015-7023.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fung-Leung WP, Schilham MW, Rahemtulla A, Kundig TM, Vollenweider M, et al. CD8 is needed for development of cytotoxic T cells but not helper T cells. Cell. 1991;65:443–449. doi: 10.1016/0092-8674(91)90462-8. [DOI] [PubMed] [Google Scholar]
- 39.Nurmi SM, Gahmberg CG, Fagerholm SC. 14-3-3 proteins bind both filamin and alphaLbeta2 integrin in activated T cells. Ann N Y Acad Sci. 2006;1090:318–325. doi: 10.1196/annals.1378.035. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz SE, Rosa JL, Scheffner M. Characterization of human hect domain family members and their interaction with UbcH5 and UbcH7. J Biol Chem. 1998;273:12148–12154. doi: 10.1074/jbc.273.20.12148. [DOI] [PubMed] [Google Scholar]
- 41.Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 42.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 43.Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 44.Liu YC. The E3 ubiquitin ligase Itch in T cell activation, differentiation, and tolerance. Semin Immunol. 2007;19:197–205. doi: 10.1016/j.smim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matesic LE, Haines DC, Copeland NG, Jenkins NA. Itch genetically interacts with Notch1 in a mouse autoimmune disease model. Hum Mol Genet. 2006;15:3485–3497. doi: 10.1093/hmg/ddl425. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto M, Takeda K, Joetham A, Ohnishi H, Matsuda H, et al. Essential role of Notch signaling in effector memory CD8+ T cell-mediated airway hyperresponsiveness and inflammation. J Exp Med. 2008;205:1087–1097. doi: 10.1084/jem.20072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet JL, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Hegele RA. Genomic basis of cystathioninuria (MIM 219500) revealed by multiple mutations in cystathionine gamma-lyase (CTH). Hum Genet. 2003;112:404–408. doi: 10.1007/s00439-003-0906-8. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Huff AM, Spence JD, Hegele RA. Single nucleotide polymorphism in CTH associated with variation in plasma homocysteine concentration. Clin Genet. 2004;65:483–486. doi: 10.1111/j.1399-0004.2004.00250.x. [DOI] [PubMed] [Google Scholar]
- 50.Hutchinson S, Aplin RT, Webb H, Kettle S, Timmermans J, et al. Molecular effects of homocysteine on cbEGF domain structure: insights into the pathogenesis of homocystinuria. J Mol Biol. 2005;346:833–844. doi: 10.1016/j.jmb.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 51.McLean RR, Jacques PF, Selhub J, Tucker KL, Samelson EJ, et al. Homocysteine as a predictive factor for hip fracture in older persons. N Engl J Med. 2004;350:2042–2049. doi: 10.1056/NEJMoa032739. [DOI] [PubMed] [Google Scholar]
- 52.van Meurs JB, Dhonukshe-Rutten RA, Pluijm SM, van der Klift M, de Jonge R, et al. Homocysteine levels and the risk of osteoporotic fracture. N Engl J Med. 2004;350:2033–2041. doi: 10.1056/NEJMoa032546. [DOI] [PubMed] [Google Scholar]
- 53.Hahn FM, Xuan JW, Chambers AF, Poulter CD. Human isopentenyl diphosphate: dimethylallyl diphosphate isomerase: overproduction, purification, and characterization. Arch Biochem Biophys. 1996;332:30–34. doi: 10.1006/abbi.1996.0312. [DOI] [PubMed] [Google Scholar]
- 54.Wilkin DJ, Kutsunai SY, Edwards PA. Isolation and sequence of the human farnesyl pyrophosphate synthetase cDNA. Coordinate regulation of the mRNAs for farnesyl pyrophosphate synthetase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and 3-hydroxy-3-methylglutaryl coenzyme A synthase by phorbol ester. J Biol Chem. 1990;265:4607–4614. [PubMed] [Google Scholar]
- 55.Kojro E, Gimpl G, Lammich S, Marz W, Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha -secretase ADAM 10. Proc Natl Acad Sci U S A. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 57.Osenkowski P, Ye W, Wang R, Wolfe MS, Selkoe DJ. Direct and potent regulation of gamma-secretase by its lipid microenvironment. J Biol Chem. 2008;283:22529–22540. doi: 10.1074/jbc.M801925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Canalis E. Notch signaling in osteoblasts. Sci Signal. 2008;1:pe17. doi: 10.1126/stke.117pe17. [DOI] [PubMed] [Google Scholar]
- 59.Avenell A, Richmond PR, Lean ME, Reid DM. Bone loss associated with a high fibre weight reduction diet in postmenopausal women. Eur J Clin Nutr. 1994;48:561–566. [PubMed] [Google Scholar]
- 60.Bedogni G, Mussi C, Malavolti M, Borghi A, Poli M, et al. Relationship between body composition and bone mineral content in young and elderly women. Ann Hum Biol. 2002;29:559–565. doi: 10.1080/03014460210137819. [DOI] [PubMed] [Google Scholar]
- 61.Chen Z, Lohman TG, Stini WA, Ritenbaugh C, Aickin M. Fat or lean tissue mass: which one is the major determinant of bone mineral mass in healthy postmenopausal women? J Bone Miner Res. 1997;12:144–151. doi: 10.1359/jbmr.1997.12.1.144. [DOI] [PubMed] [Google Scholar]
- 62.Choi JW, Pai SH. Bone mineral density correlates strongly with basal metabolic rate in postmenopausal women. Clin Chim Acta. 2003;333:79–84. doi: 10.1016/s0009-8981(03)00190-6. [DOI] [PubMed] [Google Scholar]
- 63.Frost HM. Obesity, and bone strength and “mass”: a tutorial based on insights from a new paradigm. Bone. 1997;21:211–214. doi: 10.1016/s8756-3282(97)00124-5. [DOI] [PubMed] [Google Scholar]
- 64.Hannan MT, Felson DT, Dawson-Hughes B, Tucker KL, Cupples LA, et al. Risk factors for longitudinal bone loss in elderly men and women: the Framingham Osteoporosis Study. J Bone Miner Res. 2000;15:710–720. doi: 10.1359/jbmr.2000.15.4.710. [DOI] [PubMed] [Google Scholar]
- 65.Karsenty G. Convergence between bone and energy homeostases: leptin regulation of bone mass. Cell Metab. 2006;4:341–348. doi: 10.1016/j.cmet.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cordes R, Schuster-Gossler K, Serth K, Gossler A. Specification of vertebral identity is coupled to Notch signalling and the segmentation clock. Development. 2004;131:1221–1233. doi: 10.1242/dev.01030. [DOI] [PubMed] [Google Scholar]
- 68.Hu G, Fearon ER. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–3048. [PubMed] [Google Scholar]
- 70.Matsuzawa S, Takayama S, Froesch BA, Zapata JM, Reed JC. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. Embo J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Susini L, Passer BJ, Amzallag-Elbaz N, Juven-Gershon T, Prieur S, et al. Siah-1 binds and regulates the function of Numb. Proc Natl Acad Sci U S A. 2001;98:15067–15072. doi: 10.1073/pnas.261571998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 73.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 74.Sainson RC, Harris AL. Hypoxia-regulated differentiation: let's step it up a Notch. Trends Mol Med. 2006;12:141–143. doi: 10.1016/j.molmed.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Forsyth S, Horvath A, Coughlin P. A review and comparison of the murine alpha1-antitrypsin and alpha1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics. 2003;81:336–345. doi: 10.1016/s0888-7543(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 76.Chao J, Miao RQ, Chen V, Chen LM, Chao L. Novel roles of kallistatin, a specific tissue kallikrein inhibitor, in vascular remodeling. Biol Chem. 2001;382:15–21. doi: 10.1515/BC.2001.003. [DOI] [PubMed] [Google Scholar]
- 77.Shen B, Hagiwara M, Yao YY, Chao L, Chao J. Salutary effect of kallistatin in salt-induced renal injury, inflammation, and fibrosis via antioxidative stress. Hypertension. 2008;51:1358–1365. doi: 10.1161/HYPERTENSIONAHA.107.108514. [DOI] [PubMed] [Google Scholar]
- 78.Hayward P, Kalmar T, Arias AM. Wnt/Notch signalling and information processing during development. Development. 2008;135:411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 79.Elvert R, Ehrhardt N, Taube M, Klingenspor M, Gailus-Durner V, et al. Metabolic phenotyping of mouse mutants in the German Mouse Clinic. Integrative Zoology. 2006;1:122–125. doi: 10.1111/j.1749-4877.2006.00026.x. [DOI] [PubMed] [Google Scholar]
- 80.Meyer CW, Elvert R, Scherag A, Ehrhardt N, Gailus-Durner V, et al. Power matters in closing the phenotyping gap. Naturwissenschaften. 2007;94:401–406. doi: 10.1007/s00114-006-0203-1. [DOI] [PubMed] [Google Scholar]
- 81.Meyer CW, Neubronner J, Rozman J, Stumm G, Osanger A, et al. Expanding the body mass range: associations between BMR and tissue morphology in wild type and mutant dwarf mice (David mice). J Comp Physiol [B] 2007;177:183–192. doi: 10.1007/s00360-006-0120-9. [DOI] [PubMed] [Google Scholar]
- 82.Dobush GR, Ankney CD, Krementz DG. The effect of apparatus, extraction time, and solvent type on lipid extractions of snow geese. Canadian Journal of Zoology. 1985;63:1917–1920. [Google Scholar]
- 83.Hoelter SM, Dalke C, Kallnik M, Becker L, Horsch M, et al. “Sighted C3H” mice–a tool for analysing the influence of vision on mouse behaviour? Front Biosci. 2008;13:5810–5823. doi: 10.2741/3118. [DOI] [PubMed] [Google Scholar]
- 84.Hrabe de Angelis M, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 85.Klempt M, Rathkolb B, Fuchs E, Hrabé de Angelis M, Wolf E, et al. Genotype-specific environmental impact on the variance of blood values in inbred and F1 hybrid mice. Mamm Genome. 2006;17:93–102. doi: 10.1007/s00335-005-0119-7. [DOI] [PubMed] [Google Scholar]
- 86.Kalaydjiev S, Franz T, Busch DH. Mouse phenotyping: immunology. In: Hrabe de Angelis M, Chambon P, Brown SD, editors. Standards of Mouse Models Phenotyping. Weinheim: Whiley-VCH Verlag; 2006. pp. 237–252. [Google Scholar]
- 87.Fuchs H, Schughart K, Wolf E, Balling R, Hrabe de Angelis M. Screening for dysmorphological abnormalities–a powerful tool to isolate new mouse mutants. Mamm Genome. 2000;11:528–530. doi: 10.1007/s003350010101. [DOI] [PubMed] [Google Scholar]
- 88.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, et al. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–550, 552–544, 556 passim. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- 89.Saeed AI, Sharov V, White J, Li J, Liang W, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 90.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.05 MB DOC)
(0.04 MB DOC)
(0.06 MB DOC)