Figure 5. Ikaros binds to the Rag locus and regulates histone acetylation.
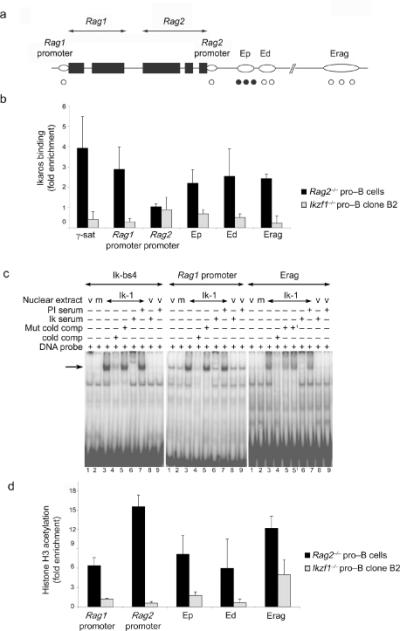
(a) Schematic representation of the Rag locus. Black boxes represent Rag1 and Rag2 exons while open ovals represent regulatory elements. Validated (•) and putative (Ο) Ikaros binding sites are indicated. (b) ChIP analysis of Ikaros binding to regulatory elements of the Rag locus. Binding was determined in Rag2-/- pro-B cells that express Ikaros and Rag1. Ikzf1-/- pro-B cells were used as negative controls. Relative enrichment of the bound DNA over input was determined by Q-PCR after normalization to Acta1. Pericentromeric g-satellite sequence (g-sat) was used as a positive control for binding of Ikaros. (c) EMSAs were performed using nuclear extracts of 293T cells transfected with the Ik-1 expression vector. Control extracts were generated from 293T cells transfected with an empty vector (lane 1) or with an Ikaros-159A DNA binding mutant (lane 2). Ik-bs4 probe contains a high affinity Ikaros binding site. Other probes spanned putative Ikaros binding sites in the Rag1 promoter and Erag enhancer. Where indicated binding reactions included competitor DNA in 50-fold molar excess or either pre-immune (PI) or anti-Ikaros antiserum (Ik serum). In lanes 5 and 51 two related mutant competitor DNAs were used. (d) ChIP analysis of histone H3 acetylation at regulatory elements of the Rag locus. Analysis was performed as described for Fig. 3b using Actb (encoding β-actin) for normalization. Error bars in panels (b) and (d) represent the standard deviation from the mean (n = 3). Data in (c) are representative of at least two independent experiments.
