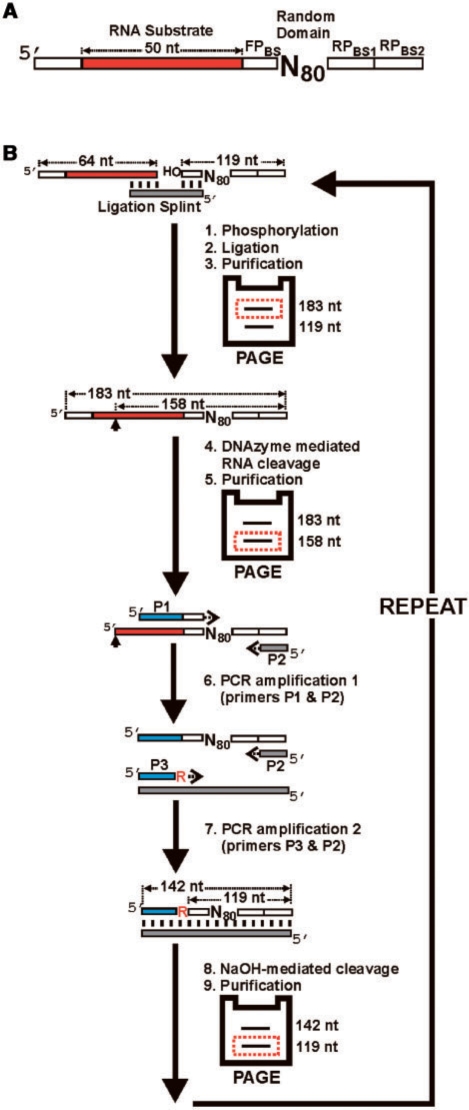
Figure 1.
In vitro selection strategy. (A) Library design. Each molecule in the library contains 80 random-sequence nucleotides (N80) as the putative DNAzyme domain, and a substrate domain composed of 50 fixed-sequence ribonucleotides (shown in red). The random domain is flanked by a 9-nt forward primer binding site (FPBS), and a 15-nt reverse-primer-binding site (RPBS). RPBS1 was used from generations 0 to 7. In generation 8, the population was split into two parallel selection experiments and different reverse primer binding sites (RPBS2) were used to minimize cross contamination during PCR. The RNA substrate is preceded by a 14-nt fixed DNA sequence intended to facilitate the separation of cleavage products during PAGE. (B) In vitro selection cycle. Each round of selection consists of steps 1–9. In step 1, the 32P-radiolabeled DNAzyme domain is 5′-phosphorylated, and then ligated to the substrate domain in step 2. The ligation product is purified by denaturing polyacrylamide gel electrophoresis (PAGE) in step 3, and incubated with the selection buffer (containing the divalent metal cofactors Mn2+ and Mg2+) to promote metal-dependent DNAzyme-mediated cleavage of the attached RNA substrate in step 4. The cleavage reaction is allowed to proceed for a designated period and stopped with the addition of the metal-chelating agent, EDTA. Although cleavage can potentially occur anywhere along the 50 ribonucleotides comprising the substrate domain, the population of DNAzymes described herein cleaved at one specific site (denoted by the arrow). The 3′-cleavage fragment(s) containing the DNAzyme domain is subsequently purified by PAGE in step 5. Two consecutive polymerase chain reactions (PCR) are used in steps 6 and 7 to amplify the cleavage fragments. The forward and reverse primers used in PCR 1 are denoted as P1 and P2, respectively. P1 contains extra nucleotides at the 5′ end (denoted in blue) that introduce a new forward priming site for the second PCR. PCR 2 uses the same reverse primer as PCR 1, but a different forward primer denoted as P3. The P3 primer contains a 3′-terminal ribonucleotide. In step 8, the double-stranded PCR product is treated with NaOH to cleave the embedded ribonucleotide, which regenerates the single-stranded DNAzyme domain. The DNAzyme domain is isolated by denaturing PAGE (step 9), and used to initiate the next round of selection.