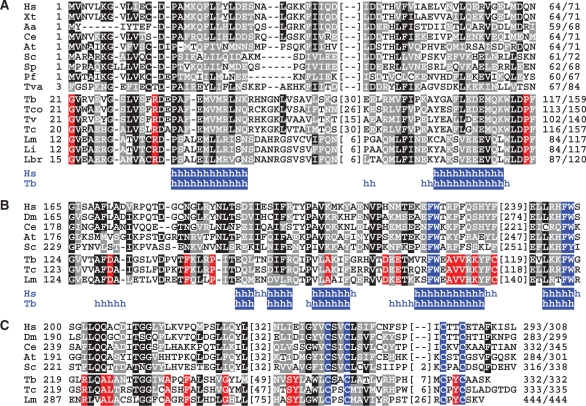
Figure 2.
Sequence alignments of TFIIH core subunits. (A) Alignment of TFB5 sequences from Homo sapiens (Hs, accession number Q6ZYL4), Xenopus tropicalis (Xt, NP_001017298), Aedes aegypti (Aa, NP_00166033), Caenorhabditis elegans (Ce, NP_497212), Arabidopsis thaliana (At, NP_172702), Saccharomyces cerevisiae (Sc, YDR079C-A), Schizosaccharomyces pombe (Sp, SPBC32F12.15), Plasmodium falciparum (Pf, PF14_0398), Trichomonas vaginalis (Tva, XP_001276879) and from the trypanosomatids T. brucei (Tb, Tb10.61.2600), Trypanosoma congolense (Tco, congo855a04.p1k_14), Trypanosoma vivax (Tv, tviv639h03.p1k_13), T. cruzi (Tc, Tc00.1047053511283.50), L. major (Lm, LmjF16.1145), Leishmania infantum (Li, LinJ16_V31190) and Leishmania braziliensis (Lbr, LbrM16_V2.1180). Positions with >50% of identical and conserved residues were shaded in black and gray, respectively. Red shading indicates positions in which trypanosomatid sequences are invariant and no conservation is present in the other sequences. Numbers of amino acids without significant similarity are specified in parentheses. Below, positions in the human and T. brucei sequences with a high probability of α-helical (h) structure are indicated. (B) Corresponding alignment of an internal domain of p62 orthologs. Shading as above except that the FW motifs are shaded in blue. Hs (P32780), Drosophila melanogaster (Dm, NP_610957), Ce (NP_499880), At (NP_175971), Sc (YDR311W), Tb (Tb11.01.1200), Tc (Tc00.1047053508851.79), Lm (LmjF36.3110). (C) Alignment of the C-terminal sequences of p34 orthologs. Shading as above except that the cysteine residues of the putative zinc finger are shaded in blue. Hs (Q13889), Dm (NP_608574), Ce (NP_499249), At (NP_564050), Sc (YPR056W), Tb (Tb11.01.7730), Tc (Tc00.1047053508707.149), Lm (LmjF32.2885).