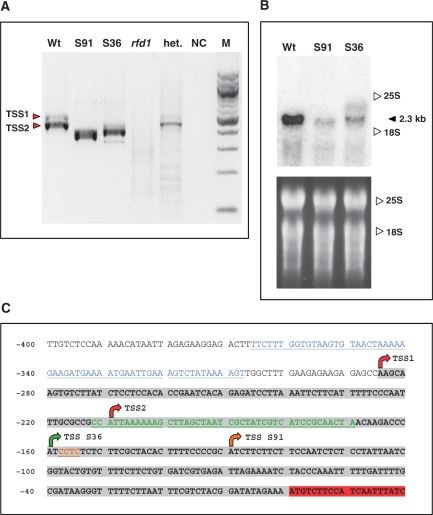
Figure 3.
(A) 5′ RACE reactions to detect AtRibA1 transcript ends in Arabidopsis wild type (Wt), SALK 036891 (S91), SALK 094736 (S36) and homozygous rfd1 seedlings (rfd1). Analysis of a heterozygous rfd1 plant (het.) and a nontemplate control (NC) are included. The DNA marker lane (M) contains a 100-bp DNA ladder (New England Biolabs, Ipswich). Red arrows depict the two major transcription start sites TSS1 and TSS2 detectable in A. thaliana wild type. (B) Northern blot of A. thaliana wild type and SALK lines 036891 and 094736, respectively. The filter was hybridized to an AtRibA1 cDNA fragment. 25S rRNA and 18S rRNA positions are indicated. The lower panel depicts the ethidium bromide stain of RNA before blotting. (C) Overview of the AtRibA1 upstream sequence with transcription start sites mapped by 5′ RACE (arrows) and T-DNA insertion sites. The 5′ UTR (highlighted in gray) in wild-type RNA results from transcription starting at TSS1 and TSS2 (red arrows). Altered transcript 5′ ends in the two SALK lines are designated TSS S36 and TSS S91. Insertions of T-DNA resulted in losses of genomic DNA stretches (colored residues) of different lengths. Insertions of pWA5 T-DNA in rfd1 (blue) and pROK2 in SALK 094736 (green) and 036891 (brown), respectively, are indicated. Nucleotide numbers are given as distances from the AtRibA1 start codon at the left margin.