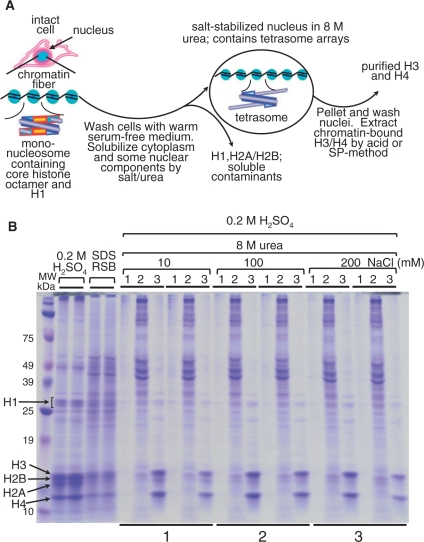
Figure 1.
(A) Flow diagram of salt-urea H3/H4 isolation from whole, undisturbed 1470.2 cells. The salt-urea treatment rapidly dissolves the cell cytoplasm and certain chromatin components, such as H1 and H2A/H2B histones. During this process, the nuclear envelope containing tetrameric H3/H4 histones and DNA (tetrasomes) is relatively well preserved and nuclei do not clump together. This phenomenon is consistently seen using a variety of salts (sodium phosphate, NaCl, KCl, etc.), suggesting that nuclei stabilization is not salt-specific. (B) High yield recovery of H3 and H4 from intact cells by using high urea and salt concentrations. Duplicate samples (bottom underlines 1, 2 and 3) were treated with 8-M urea at increasing concentration of NaCl (10, 100 and 200 mM, as indicated at the top). The histones were sulfuric-acid extracted, TCA precipitated, and resuspended in 80 μl of denaturing reducing sample buffer (RSB) for analysis on 12.5% SDS–PAGE and stained with Coomassie blue. Duplicate histone samples, extracted with H2SO4 and 2× reducing sample buffer (RSB), are shown for comparison (four lanes next to marker lane). The RSB sample contains all cellular proteins and is used as a control for histone yield and purity. With the exception of the RSB samples (which were loaded directly), the extracted histones were TCA precipitated and resuspended in 80 µl of RSB for gel loading. Five microliters of each sample were loaded per well. Indicated at the top of the gel are: lane 1, H2SO4-insoluble cell debris after H3/H4 extraction; lane 2, cell cytoplasm and some nuclear proteins solubilized in 8-M urea at increasing NaCl concentrations (indicated at the top of the gel); lane 3, H2SO4-extracted H3/H4 after salt-urea treatment.