Abstract
PCNA has no intrinsic enzymatic function, but functions as a sliding platform to mediate protein interactions with the DNA strand. Many proteins interact with PCNA through a small conserved motif with consensus QxxLxxFF. This work uses S. pombe and human cells to analyse the function of PCNA-binding peptides. Interacting peptides were identified using two-hybrid screening; one (pep102) binds directly to a physiologically relevant site on PCNA. The EGFP-pep102 over-expression phenotype is consistent with competitive blocking of PCNA-protein interactions. Various PCNA-binding peptides were all shown to inhibit PCNA function by competitive binding in both human and S. pombe cells as EGFP fusion proteins. The action of a p21(WAF1/Cip1)-derived peptide was complicated by the presence of additional functional domains and possible post-translational modification. The activity of pep102 was hampered by low expression in both model systems. The peptide derived from rational design (con1) was stable, highly active in inhibiting PCNA function both S. pombe and human cells and showed a high affinity for PCNA both in vitro and in vivo. These results validate the use of functional screening in yeast to identify peptide aptamers that are functional in mammalian cells; such aptamers provide excellent leads for small molecule anti-proliferative therapies.
Keywords: PCNA, peptides, interaction, screening
Introduction
Cellular processes are regulated by complex patterns of protein-protein interactions. The perturbation of these interactions by the expression of transdominant suppressors can provide useful tools for the analysis of these regulatory processes (Kamb & Caponigro, 2001). Such suppressors can take many forms, such as dominant negative forms of proteins, anti-sense RNA and nucleic acid aptamers. The use of peptide aptamers is rapidly becoming accepted as one of the most useful forms of transdominant suppressor technology. Such aptamers are capable of high affinity interactions with their targets and can show highly specific target recognition. The strength of peptide aptamers lies in the use of genetic techniques for their identification and analysis. A pre-determined target or gene can be screened using a reverse genetic approach to identify small peptide sequences that interact specifically with the target. Several methodologies are available for this, including phage display and various forms of two-hybrid screening. Once interactors have been identified, assays can be undertaken to determine whether binding of the peptide ligand to the protein affects protein function, either in vitro or in vivo. Transdominant suppressors of specific protein function are of particular value in the study of cell types, such as mammalian cells, which are relatively intractable to traditional genetic approaches (Xu et al., 2001b). Although techniques such as RNAi can be used to lower global protein levels, peptide aptamers have the advantage that the functional consequences of specific protein-protein interactions can be targeted.
In the work described here, screening was undertaken for peptides that interact with Proliferating Cell Nuclear Antigen (PCNA). This is a ubiquitous eukaryotic protein which is essential for DNA replication, various forms of DNA repair and in post-replicative processing (Jonsson & Hubscher, 1997; Kelman, 1997; Tsurimoto, 1999). PCNA assembles as a trimer with a toroidal shape which is capable of encircling double stranded DNA. In this way, it enhances polymerase processivity by tethering the polymerase complex to the target DNA. Many of the proteins which interact directly with PCNA are involved in the mechanics of DNA replication and repair including Replication Factor C (RFC), DNA polymerase δ, DNA ligase 1, Fen1, XPG, MSH3 and MSH6 (Kelman & Hurwitz, 1998). PCNA also interacts with proteins involved in post-replicative processing, including DNA (cytosine-5) methyltransferase, uracil DNA glycosylase (UNG2) and chromatin assembly factor (CAF1) (Chuang et al., 1997; Moggs et al., 2000; Otterlei et al., 1999) and with cell cycle regulatory proteins such as Gadd45 and p21(WAF1/Cip1) (Chen et al., 1995; Flores-Rozas et al., 1994; Hall et al., 1995; Smith et al., 1994; Waga et al., 1994; Warbrick et al., 1995). The interaction of PCNA with these regulatory proteins suggests that it may link the DNA damage response with the regulation of DNA replication and repair.
A clue to the mechanism by which this regulation may function is the observation that many, though not all, of these PCNA-binding proteins interact through a small conserved motif which binds to a common site on PCNA (Warbrick, 2000). These results indicate that PCNA plays a central role in co-ordinating cell cycle events with DNA replication and repair through the consecutive, regulated binding of proteins to PCNA through a common interaction site. This interaction site, therefore, provides a particularly good model target for designing therapeutic agents to treat hyperproliferative diseases such as cancer. p21-derived agents have been shown to block replication and cell cycle progression in vitro and in cultured cells (Cayrol et al., 1998; Mattock et al., 2001a; Mattock et al., 2001b; Warbrick et al., 1995). Peptides and small molecules which can specifically disrupt protein-PCNA interactions present potentially useful tools in the analysis of the functional roles of these interactions.
In order to identify peptides which bound to sites on PCNA which are exposed in vivo, we have successfully used yeast two-hybrid techniques for screening. A particular peptide was chosen for further analysis which shows sequence specific binding to PCNA from a range of species. We show that this peptide can interact with PCNA in both human and S. pombe cells with functional consequences for both types of cell resulting in the inhibition of proliferation. We have compared the activity of this peptide with that of PCNA-binding peptides derived from the human protein p21(WAF1/Cip1) and with a rationally designed PCNA-binding peptide. The comparison of the activity of these peptides has important implications in the use of yeast as a model organism in which to screen for biologically active peptides that have activity in inhibiting conserved protein pathways such as that involving the interaction with PCNA. The observation that these peptides are active in both human and S. pombe cells validates the use of functional screening in yeast to identify peptide aptamers that are functional in mammalian cells.
Results
Identification of PCNA-binding peptides by two-hybrid screening of a peptide library
Two-hybrid screens using either human or S. pombe PCNA were used to identify novel PCNA-binding peptide sequences. The library used expresses a 16 amino acid random sequence at the C-terminus of the transcriptional activation domain of Gal4 and contains approximately 1 × 107 independent clones (Yang et al., 1995). For S. pombe PCNA, one positive clone containing plasmid p7s was identified from approximately 2 × 107 transformants. For human PCNA, three positive clones containing plasmids p102h, p103h and p104h were identified from approximately 4 × 107 transformants. The four plasmids identified each showed an interaction with human, S. pombe and Drosophila melanogaster PCNA in two-hybrid tests. They did not interact with unrelated control proteins, or activate transcription alone. DNA sequencing showed that the inserts in plasmids p102h, p104h and p7s were identical while the insert in p103h was distinct.
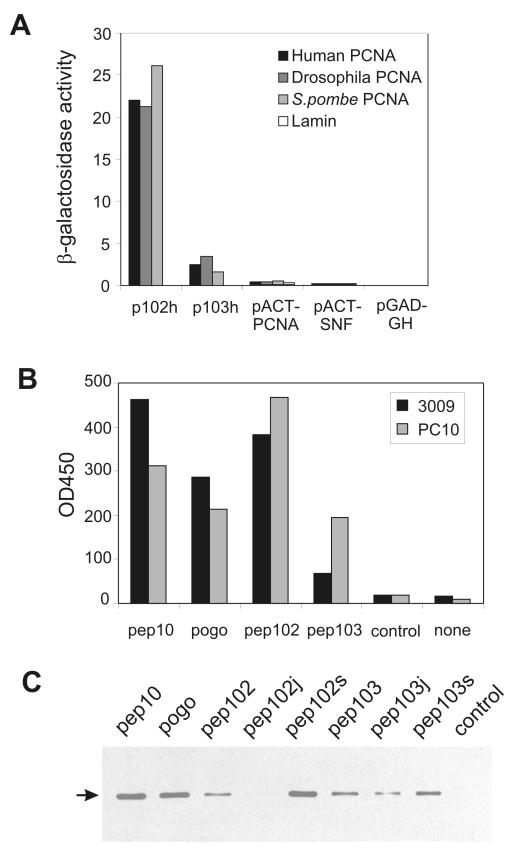
Plasmids p102h and p103h were analysed by assessing the ability of plasmid pairs to activate the reporter LacZ as measured by a β-galactosidase assay (Figure 1A). p102h shows an exceptionally strong interaction with PCNA from all three species, which is at least an order of magnitude greater than the interaction of PCNA with itself in this system. The peptides encoded by these plasmids did not share homology to the conserved PCNA-interaction domain defined by the PCNA-binding consensus QxxLxxFF or to other known protein sequence (see Discussion).
Figure 1. In vitro and in vivo analysis of peptide-PCNA interactions.
Panel A: Results of β-galactosidase assays on two-hybrid transformants. p102h and p103h are as described in the text; pACT-PCNA expresses full length S. pombe PCNA as a fusion with Gal4ACT pACT-SNF expresses yeast SNF4 as a control and pGAD-GH is the library vector. These were tested against PCNA from various species expressed as fusions with Gal4AS. “Lamin” was used as a negative control. Panel B: ELISA assays of human PCNA binding to peptides. Peptides were immobilised on streptavidin-coated multiwell plates through an N-linked biotin moiety (as indicated) and binding of purified human PCNA detected using two anti-PCNA antibodies, 3009 or PC10. Control indicates an unrelated peptide and none indicates that no peptide was used for affinity capture. Panel C: Affinity capture experiments were carried out from HeLa cell extracts with peptides as shown; bound PCNA was detected by Western blot as indicated by the arrow.
Synthetic peptides were synthesised for further analysis (see Experimental Procedures). Pep102 corresponds to the sequence encoded by plasmids p102h, p104h and p7s. Pep103 corresponds to that in plasmid p103h. Pep10 is a PCNA-binding peptide derived from human p21(WAF1/Cip1) protein as previously described (Mattock et al., 2001a; Warbrick et al., 1995). Pogo is a PCNA-binding peptide derived from the Drosophila Pogo transposase (Warbrick et al., 1998). In initial experiments, an ELISA-type assay was used in which the relative amounts of purified human PCNA interacting with immobilised peptides was assessed by antibody detection. The results show that pep102 and pep103 can bind directly to PCNA (Figure 1B). Pep102 bound to PCNA with a high affinity, which was similar to that shown by peptide pep10. Pep103 also showed significant binding, but with a lower affinity. Comparison of results obtained using two anti-PCNA antibodies showed that PCNA bound to pep102 and pep103 showed a stronger reaction with PC10 rather than 3009, while the converse was true with pep10 and pogo. To avoid problems involving the antibody detection of PCNA, peptides were bound to streptavidin-agarose beads and bound PCNA detected by SDS-PAGE followed by Western blotting in subsequent experiments. Using this method, peptides were tested for binding to PCNA in cell extracts from HeLa cells (Figure 1C) and from a variety of species (data not shown). Pep10, pogo and pep102 bound to PCNA efficiently from all the extracts tested, suggesting that the interaction is evolutionarily conserved. In contrast, pep103 bound PCNA from human extracts, but bound PCNA from the other species tested much less efficiently (data not shown).
To investigate the specificity of the interaction of pep102 and pep103 with PCNA, additional variants of these peptides were analysed. These included shorter peptides representing the 16 amino acids sequence encoded by the library oligonucleotide with no flanking sequences (“s”) and “jumbled” peptides in which these 16 amino acids were randomised (“j”; see Materials and Methods for details of peptide sequences). The ability of these peptides to bind to PCNA from HeLa extracts is shown in Figure 1C. The 16 amino acid pep102s peptide showed a strong interaction with PCNA, while jumbling of the sequence dramatically reduced PCNA-binding, indicating that the interaction is sequence-specific. In contrast, significant amounts of PCNA bound to the jumbled 103 peptide (pep103j), indicating that the interaction of pep103 with PCNA is likely to be non-specific. The pep103 sequence was not carried forward for further analysis.
The novel peptide pep102 shares a binding site with physiological PCNA interaction domains
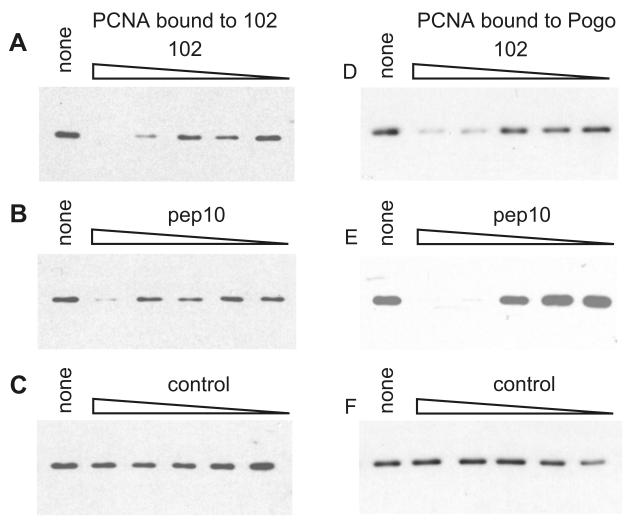
Peptide competition experiments were carried out to investigate whether pep10 and pep102 share a binding site on PCNA. The ability of bead-linked peptides to bind to PCNA in HeLa extracts was tested in the presence of non-biotinylated peptides. With pep102 bound to beads, free pep102 and free pep10 effectively competed for binding to PCNA (Figure 2A, B). Free pep10 has previously been shown to compete with the pogo peptide for binding to human PCNA (Warbrick et al., 1998). Although free pep102 does not compete for PCNA binding to pep10 (data not shown), both pep10 and pep102 compete for binding to pogo at similar concentrations (Figures 2D, E). Taken together, the evidence from these competition experiments suggests that pep10 and pep102 bind to overlapping regions of human PCNA, with pep10 showing the higher affinity. This is interesting, given the lack of any apparent sequence similarity between pep10 and pep102. However, a possibility which cannot be excluded is that pep102 binds to a distinct site on PCNA and induces a conformational change that prevents the binding of either the pep10 or pogo peptides.
Figure 2. Peptides share a common binding site on human PCNA.
The ability of bead-conjugated peptides to bind to PCNA in HeLa cell extracts was tested in the presence of varying concentrations of free, unbiotinylated peptides as described in Materials and Methods. The relative amounts of bound PCNA are shown by Western blot analysis. In panels A, B and C the ability of PCNA in HeLa cell extracts to bind to 102 conjugated beads was tested, in the presence of various competing peptides. In panels D, E and F, the ability of PCNA to bind to pogo-conjugated peptide beads was tested. The addition of an unrelated peptide (control) did not affect the biotinylated peptide-PCNA interaction.
Pep102 interacts with human PCNA in vivo
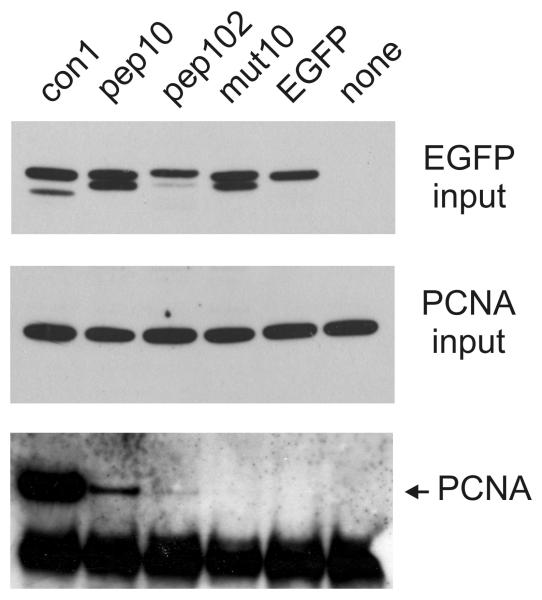
To analyse the ability of the pep102 sequence to interact with human PCNA in vivo, it was expressed as a C-terminal fusion with EGFP in human cells (EGFP-pep102). This was compared with an EGFP-pep10 fusion and with EGFP fused to the mut10 sequence, which is a mutated version of pep10 region that cannot bind to PCNA (Mattock et al., 2001a; Mattock et al., 2001b). The EGFP-peptide fusions showed varying levels of expression, with EGFP-mut10 expressed at a higher level than EGFP alone and EGFP-102 expressed at a lower level (Figure 3 and data not shown). The reason for this is not clear. EGFP-pep10 and EGFP-mut10 have previously been shown to have a predominantly nuclear localisation (Mattock et al., 2001b). EGFP-102 and EGFP-con1 are also localised to the nucleus (data not shown). Immunoprecipitation experiments using a polyclonal anti-EGFP antibody showed that EGFP and the EGFP-peptide fusion proteins could be successfully immunoprecipitated (data not shown). As observed previously, PCNA was co-immunoprecipitated with EGFP-pep10, though not with EGFP-mut10 or with EGFP alone (Mattock et al., 2001b). The con1 sequence is a consensus PCNA-binding peptide derived from rational design which has a high affinity with human PCNA (Zheleva et al., 2000). The structure of con1 (also known as the PL peptide) has recently been solved by x-ray crystallography (Kontopidis et al., 2005). Far more PCNA was co-immunoprecipitated with EGFP-con1 than with EGFP-pep10, which is surprising, as their affinities for PCNA in vitro have been shown to be similar (Zheleva et al., 2000). Significant amounts of PCNA were co-immunoprecipitated with EGFP-pep102, indicating that human PCNA interacts with this peptide sequence in vivo (Figure 3, lower panel).
Figure 3. Western blot analysis of EGFP-peptide constructs expressed in vivo.
Peptide-EGFP fusion proteins were expressed in human U2OS cells and cell lysates prepared as described. Immunoprecipitation was carried out with the polyclonal anti-EGFP antibody Ab290 (AbCam) and the precipitated proteins analysed by SDS-PAGE and Western blot. “EGFP” indicates that EGFP was expressed from the vector with no additional fusion sequence; “none” indicates transfection with an unrelated plasmid. Other EGFP fusions with peptides are as shown. The amounts of plasmid DNA used for transfection of a 10cm dish were as follows: pEGFP-con1: 10μg; pEGFP-pep10: 10μg; pEGFP-102: 30μg; pEGFP-mut10: 2.5μg; pEGFP: 10μg. In the upper panel, input levels of pEGFP fusion proteins are shown in a Western blot probed with a mouse monoclonal anti-EGFP antibody (Roche). The middle panel shows input levels of PCNA. The lower panel shows Western blot analysis of PCNA co-immuoprecipitated with pEGFP fusion proteins. The lower band in this panel is cross-reactivity of the antibody light chain.
Peptide 102 induces a PCNA-dependent cdc phenotype in S. pombe
We investigated the physiological effects of inducing expression of the pep102 sequence in S. pombe strains containing integrated expression constructs. Following 12 hours induction of protein expression, levels of EGFP-pep102 were strongly induced and increased following further growth as shown by Western blotting (data not shown).
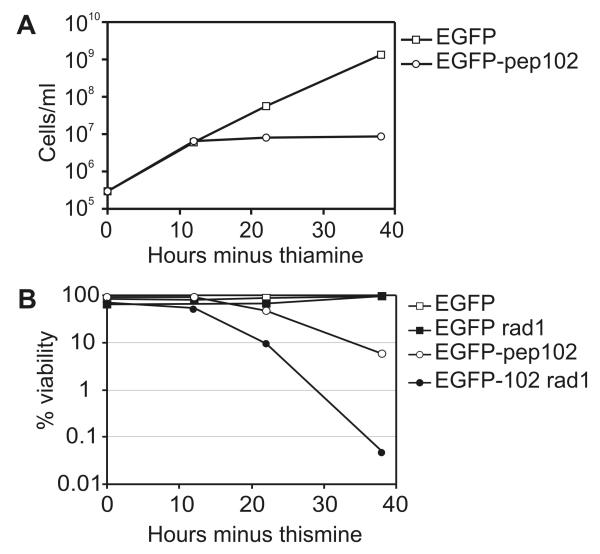
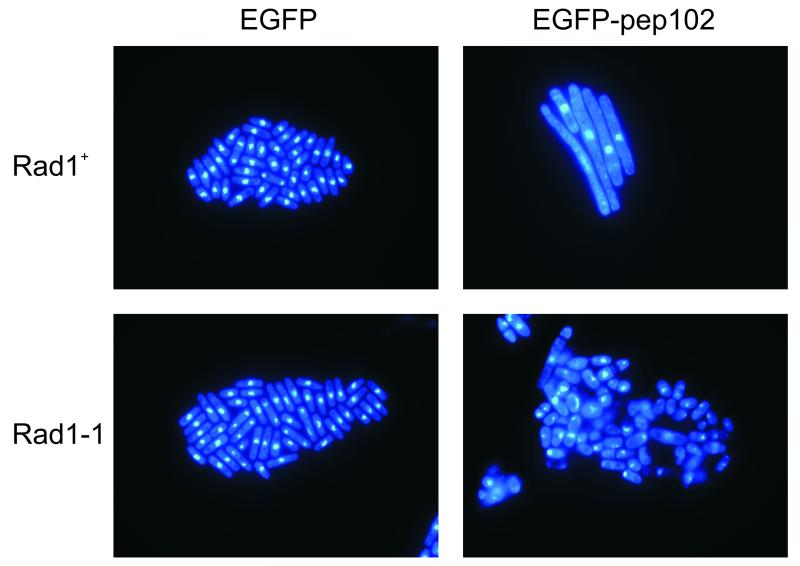
Analysis of cell number showed that while EGFP expression had no detectable effect, proliferation was inhibited by expression of EGFP-pep102, so that little or no increase in cell number occurred following 10 hours induction (Figure 4A). Cell viability also began to decrease following induction of EGFP-pep102 (Figure 4B). Cells expressing EGFP-pep102 were highly elongated with a single nucleus, which is characteristic of a cell cycle-dependent arrest (Figure 5). Following EGFP-pep102 induction in a rad1 background, cells lost viability very quickly compared to rad1+ cells (Figure 4B). These cells were not elongated, but showed a range of aberrant phenotypes characteristic of mitotic catastrophe (Figure 5). Rad1 belongs to the family of checkpoint rad proteins which function in a feedback control which inhibits cell cycle progression in response to both DNA damage and the completion of DNA synthesis (al-Khodairy et al., 1994). These results indicate that expression of EGFP-pep102 leads to a checkpoint-dependent arrest, which is typical of mutants that result in incomplete DNA replication such as those in cdc1+ and cdc27+ which encode subunits of polymerase δ (MacNeill et al., 1996; Reynolds et al., 2000). These results therefore suggest that the interaction of EGFP-pep102 with PCNA results in a block to DNA replication.
Figure 4. The effects of EGFP-pep102 expression on cell number and viability in S. pombe.
Protein expression was induced in S. pombe cells containing integrated constructs expressing either EGFP or EGFP-pep102 by inoculating log phase cells into thiamine-free medium. Cell numbers (A) and viability (B) were determined and the results of a representative experiment shown. “rad1” indicates the transformants had a rad1-1 genetic background.
Figure 5. Expression of EGFP-pep102 in S. pombe results in a checkpoint-dependent cell cycle arrest.
Following induction of protein expression, cells expressing EGFP or EGFP-pep102 were harvested, fixed and stained with DAPI as described. The results of expressing EGFP-pep102 are shown (right column) compared to those of EGFP alone (left column) either in a rad1+ (upper row) or a rad1-1 genetic background (lower row).
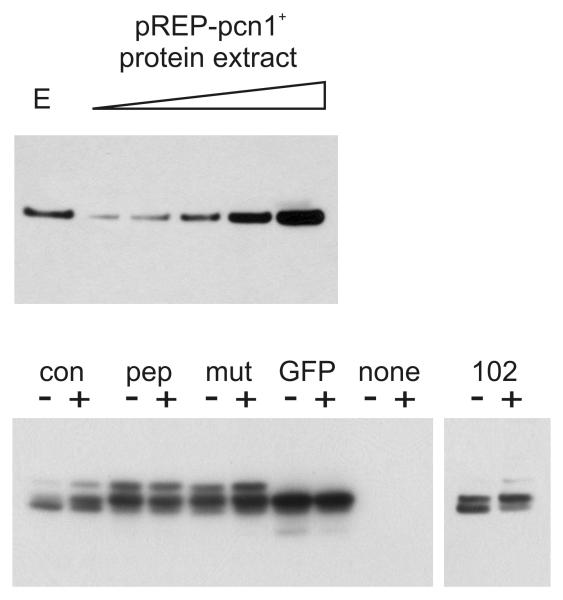
To confirm that the effect of EGFP-pep102 expression was PCNA-dependent, a plasmid expressing S. pombe PCNA encoded by pcn1+ was introduced into EGFP- and EGFP-pep102 expressing strains. The phenotype of these transformants is complicated by the observation that high levels of expression of S. pombe PCNA leads to a cell cycle delay and a slightly reduced growth rate (Waseem et al., 1992). However, the effect of high-level PCNA expression in these strains could be assessed by examining colony size following the streaking out of transformants upon minimal medium plates lacking thiamine. A comparison was made of the growth of either rad1+ or rad1-1 strains stably expressing either EGFP or EGFP-pep102 under the control of the nmt42 promoter. A summary of these results is shown in Table 1. Expression of PCNA effectively suppressed the phenotype caused by the expression of EGFP-pep102 in both rad1+ and rad1- genetic backgrounds. The level of PCNA when over-expressed in this system is between 200 and 300 times that of the endogenous protein (Figure 6). These observations using PCNA co-expression are highly suggestive that the phenotypic effects of EGFP-102 are a direct result of its interaction with PCNA.
Table 1. High-level expression of S. pombe PCNA can suppress the cell cycle lethality caused by expression of EGFP-pep102.
| Strain | Transformed plasmid |
|
|---|---|---|
| pREP1 | pREP1-pcn1+ | |
| (EGFP) | Normal growth | Slightly reduced growth |
| (EGFP) rad1-1 | Normal growth | Slightly reduced growth |
| (EGFP-pep102) | Tiny colonies | Normal growth |
| (EGFP-pep102) rad1-1 | No colonies | Slightly reduced growth |
Figure 6. Levels of pEGFP-fusion proteins and PCNA expressed in S. pombe.
Upper panel shows a comparison of endogenous PCNA with levels in dilutions of protein extracts from cells over-expressing PCNA. Protein expression was induced for 24 hours in cells containing either vector (pREP1) or pREP1-pcn1+, which expresses S. pombe PCNA under the control of the nmt promoter. Soluble protein extracts were prepared and analysed by SDS-PAGE and Western blot analysis using the anti-PCNA antibody PC10. “E” indicates endogenous PCNA present in the vector-only transformants. In the remaining five lanes, increasing amounts of protein extract from pREP1-pcn1+ transformants was loaded at dilutions of 1 in 500, 1 in 400, 1 in 300, 1 in 200 and 1 in 100. The lower panel shows levels of EGFP and EGFP-peptide fusion proteins expressed with (+) and without (−) co-expressed PCNA. Protein expression (as above) was induced for 24 hours. The panel showing EGFP-102 expression has been exposed approximately 8 times longer than the main panel.
We have already shown that the binding site of pep102 is shared with that of peptides containing the conserved PCNA-binding motif. Proteins containing this motif are involved in a wide range of DNA related processes, including DNA replication, repair, and post-replicative processing (Warbrick, 2000). We therefore investigated whether EGFP-pep102 inhibited DNA repair in S. pombe. Table 2 shows that EGFP-pep102-expressing cells are significantly more sensitive to UV compared to the control suggesting that pep102 is competing with cellular proteins involved in DNA repair.
Table 2. EGFP-pep102 expression results in UV sensitivity*.
| Minus UV | Plus UV | % UV viability | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| EGFP | EGFP- pep102 |
EGFP | EGFP- pep102 |
EGFP | EGFP- pep102 |
|
| 12 hours | 81% | 91% | 52% | 45% | 64% | 49% |
| 22 hours | 86% | 47% | 63% | <1% | 73% | <1% |
“UV viability” was calculated as (% viability plus UV)/(% viability minus UV)×100
A comparison of PCNA-binding peptides identified using various approaches
While analysing the expression of the pep102 sequence in S. pombe, it was observed that EGFP expression from pREP42-EGFPC was very low compared to EGFP-pep102 using the same plasmid vector (data not shown). To address this anomaly, a new set of vectors was created to express C-terminal EGFP fusion proteins in S. pombe. These contain various forms of the nmt promoter, an optimised Kozac consensus sequence and an improved polylinker sequence (see Materials and Methods). These vectors are p4xG, p42xG and p82xG which use the nmt high, medium and low level attenuated promoters and have the ura4+ sequence for selection. Sequences encoding pep10, mut10, pep102 and con1 were cloned into all these vectors. Western blot analysis shows that these EGFP-peptide fusion proteins are all expressed at similar levels in the p4xG vector following 24 hours induction of expression, with the exception of EGFP-102, which, as in human cells, is expressed at a lower steady state level (Figure 6).
Transformed cells were streaked upon selective medium under inducing conditions and colony formation assessed. EGFP-con1 expression had the strongest effect, with growth significantly impaired. When the peptides were expressed most strongly using the p4xG vector, EGFP-con1 tranformants showed little or no colony formation and EGFP-pep102 transformants showed only very small colonies. These transformants both showed a highly elongated cdc phenotype with EGFP-con1-expressing cells showing a longer average length. However, EGFP-pep10- and EGFP-mut10-expressing transformants could still form colonies when grown under inducing conditions, though their growth was slightly impaired, and they did not have an elongated phenotype. Colony formation of p4xG-pep102 and p4xG-con1 transformants could be partially restored by co-transformation with pRep1-pcn1+ as described above, indicating that the cdc phenotype resulting from con1 expression was due to its interaction with PCNA, as was shown for EGFP-pep102. Co-expression of PCNA had no effect upon the expression levels of the peptide fusions as assessed by Western blot (Figure 6).
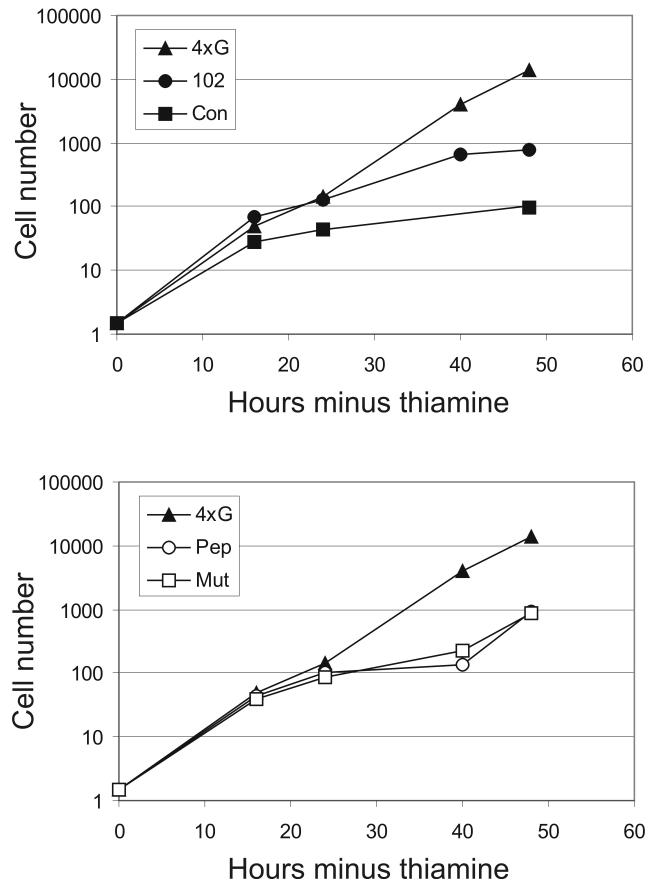
The growth of these transformants was monitored in liquid culture following induction of the EGFP-peptide fusion expressed from the p4xG plasmid vector (Figure 7). EGFP-con1 showed a very strong effect upon proliferation and significantly elongated cells were observed even at the 16-hour time point. Although no cell elongation was seen in the pEGFP-pep10 and pEGFP-mut10 transformants, a significant effect upon proliferation was observed (Figure 7; lower panel). Following 40 hours incubation in the absence of thiamine, these transformants began to escape from their arrest. This is consistent with the observation that these transformants could form colonies when grown on solid medium. pep10 and mut10 peptides showed a completely different effect compared to the other PCNA-binding peptides. Since the effects of pep10 and mut10 were indistinguishable, their activities are most likely due to the cyclin binding motif KRLIFS which is present in both peptides.
Figure 7. The effects of EGFP-peptide expression upon cell number in S. pombe.
Following induction of protein expression by inoculation into medium lacking thiamine, cell numbers were determined for transformants expressing various EGFP-peptide fusion proteins as shown. 4xG indicates EGFP expression with no fusion from the vector. The results of a representative experiment are shown.
FACS analysis was used to further investigate the effect of these peptides upon the cell cycle. Although those cells expressing the highest levels of EGFP could not be included in the analysis due to bleed-through of the very strong EFGP fluorescence between channels, no significant G1 population could be seen in any of the EGFP-peptide expressing transformants following up to 36 hours incubation without thiamine. Following 40 hours incubation, a small G1 population could be seen appearing in the pEGFP-pep10 and pEGFP-mut10 transformants (data not shown). This is consistent with an escape from G2 arrest and the concomitant increase in cell number seen at that time point. Thus the effect of pep10 and mut10 is to arrest the cells with a 2N DNA content, but without showing a cdc phenotype, which is a suggestive of an arrest in early mitosis rather than at the G2-M boundary such as that seen in the pEGFP-pep102 and pEGFP-con1 transformants.
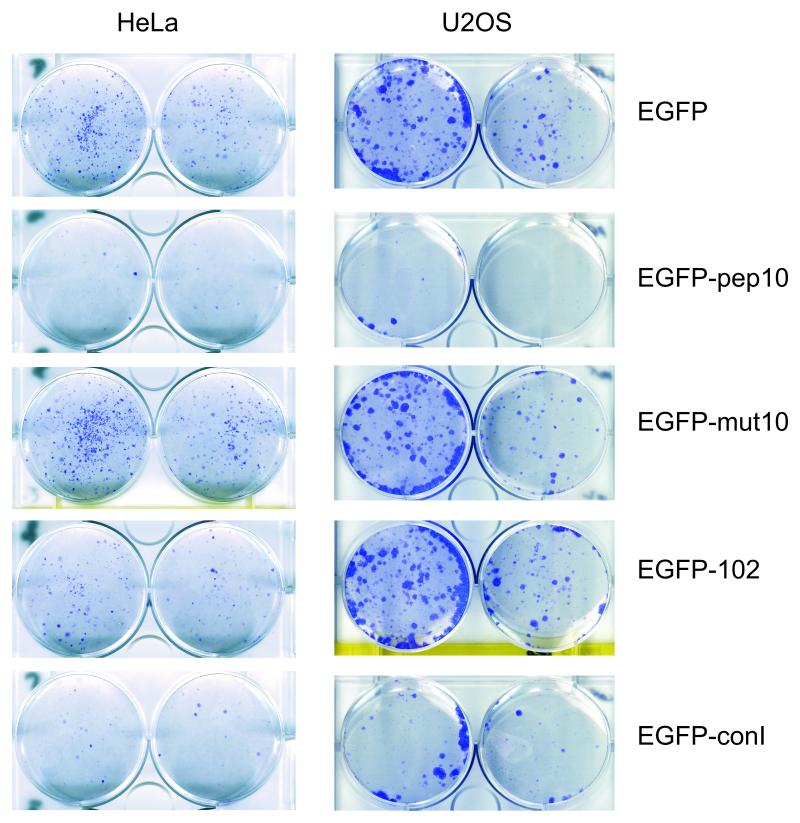
In human cells, it has previously been described that expression of the pep10 sequence inhibits proliferation in a range of cell lines. Mutant variants of this sequence which could not interact with PCNA did not have this effect, indicating that the effect is PCNA dependent (Mattock et al., 2001b). Similar experiments were carried here out to compare the effect of EGFP-pep102 and EGFP-con1 with EGFP-pep10 and EGFP-mut10 in clongenic assays (Figure 8). EGFP-pep10 has a very strong anti-proliferative effect compared to EGFP alone or EGFP-mut10 in both U2OS and HeLa cells. EGFP-con1 also has a very strong effect which is consistent with its high affinity interaction with PCNA in vivo and its strong, PCNA-dependent effect upon the cell cycle in S. pombe. However, pep102 shows only a limited effect in HeLa cells and no distinguishable effect in U2OS cells. One possible reason for this is the relatively low level of expression of the EGFP-pep102 fusion (data not shown and Figure 3).
Figure 8. EGFP-peptide fusions inhibit colony formation in human tumour cell lines.
Clonogenic assays were carried out to assess the effects of EGFP-peptide fusions – for details see Materials and Methods. Transfected cells were grown for 10 to 20 days under selection before fixation and staining with Giemsa. For each pair of wells, the left hand wells were seeded with twice as many cells as the right. Representative results are shown in each case for HeLa (cervical cancer) and for U2OS (osteosarcoma) cell lines.
Discussion
In many basic cellular pathways, yeast and human cells show a very high level of conservation at the protein structural level. For example, many of the proteins and pathways involved in DNA replication and repair were first elucidated in yeast. There are many examples of functional conservation, the first example of which was the discovery that the human Cdk2 gene could complement cdc2 mutations in Schizosaccharomyces pombe (Lee & Nurse, 1987). The human PCNA gene can complement a Pcn1 deletion in S. pombe (Waseem et al., 1992).
To analyse peptides which have activity in both a model system and in human cells we chose to use S. pombe as the model and used the two-hybrid system to screen a peptide fusion library against both human and S. pombe PCNA. Screening peptide libraries using the two-hybrid system has several advantages: protein interactions are assessed in a cellular environment and screening can be highly sensitive. One peptide sequence (pep102) was identified as interacting wit both human and S. pombe PCNA and was later shown to have affinity for PCNA from every species tested. The sequence of this peptide shows no detectable homology to previously characterised PCNA-binding domains and no significant homologies were found by searching protein sequence databases. A number of peptide sequences which bind to PCNA in vitro have been identified: none of these showed significant homology with the peptide identified here (Xu et al., 2001a).
Despite the lack of sequence homology, peptide competition experiments show that the binding site of pep102 on PCNA overlaps with the interaction domain of p21(WAF1/Cip1). pep102, therefore, binds to PCNA at a physiologically relevant interaction site, which supports evidence that the two-hybrid system offers a close parallel to physiological conditions. The hydrophobic residues within pep102 may fit into the “pocket” which interacts with the MxxFY motif in the p21 PCNA-binding motif (Gulbis et al., 1996).
When expressed as an EGFP miniprotein, the pep102 sequence has a functional activity in vivo which is consistent with its ability to inhibit protein-PCNA interactions by competitive binding. Expression of pep102 in S. pombe inhibits the cell cycle in a checkpoint-dependent manner and also results in sensitivity to DNA damage. These phenotypes can be abrogated by high-level PCNA expression, suggesting that they are a direct result of the PCNA-peptide interaction. Many proteins required for DNA replication and repair, including Fen1, DNA ligase I and polymerase δ, interact with PCNA through the conserved PCNA binding motif. The results shown here are consistent with the pep102 preventing PCNA interactions of these proteins. The analysis of pep102 function in human cell lines have proved more difficult due to very low expression levels, the reason for which is not clear. However, in HeLa cells, expression of pep102 clearly inhibits cell proliferation in clonogenic assays which is consistent with its activity as a PCNA-binding peptide (see below).
Other PCNA-binding peptides have previously been described: the PCNA-binding region of the human p21(WAF1/Cip1) protein has been extensively characterised and the structure of this peptide co-crystallised with human PCNA has been solved (Gulbis et al., 1996). However this region also contains a bi-partite nuclear localisation sequence and a cyclin-binding motif (KRLIFS) which can inhibit the activity of CDK-cyclin complexes to specific substrates by competitively inhibiting the cyclin-substrate interaction (Adams et al., 1996; Zheleva et al., 2002). A 20 amino-acid peptide derived from p21(WAF1/Cip1) encompassing all three functional domains (pep10) has been shown to inhibit proliferation in human cells lines specifically by its interaction with PCNA. This was demonstrated by comparing its activity with a mutant form of the peptide sequence in which specific PCNA-interacting residues were mutated leaving the cyclin binding activity of the peptide intact (Mattock et al., 2001b). The con1 PCNA-binding peptide was the result of rational design aimed at producing a peptide which would share an interaction site with the pep10 sequence, but which would not have the cyclin-binding activity of pep10. This 16 amino peptide interacts with PCNA with a similar affinity to that shown by pep10 in vitro and has a similar ability to inhibit SV40 replication in vitro (Zheleva et al., 2000).
In order to compare the effects of peptides derived from protein sequences, peptides derived from screening and rationally designed peptides, we expressed pep10, pep102 and con1 as EGFP miniproteins and compared their activity with the pep10 variant mut10 and EGFP alone in both S. pombe and human cells lines. We already knew that, in vitro, pep10 and con1 show a similar affinity to PCNA, while that of pep102 sequence was significantly lower (Kontopidis et al., 2005). Although pep102 showed a significant PCNA-dependent effect in inhibiting the cell cycle in S. pombe, the effect of con1 expression was even stronger as judged by timing of cell length increase, which was concomitant with the inhibition of proliferation, and also the stringency of the cell cycle block. In human cell lines, the analysis of pep102 was complicated by very low expression levels of the pep102-EGFP miniprotein. However, an effect upon cell proliferation could be seen. Consistently with its high affinity for PCNA and its strong effect upon the cell cycle in S. pombe, con1 effectively inhibited cell proliferation with a similar efficiency to that shown by pep10 in human cell lines. Given that the con1 and pep10 peptides show a similar affinity to purified PCNA in vitro, it was surprising that, in vivo, EGFP-con1 appeared to interact more stably with PCNA than EGFP-pep10. One possible explanation is that pep10 may undergo post-translational modification; Thr145 and Ser146 of p21 (equivalent to residues 5 and 6 in pep10) can be phosphorylated by AKT/PKB in vitro and in vivo and Thr145 can inhibit PCNA binding (Li et al., 2002; Rossig et al., 2001).
In contrast to the pep102 and con1 peptides, the effect of pep10 in S. pombe appeared to be a consequence of its cyclin-binding motif. In human cells, results shown here and those previously published show that pep10 inhibits cell proliferation in a PCNA-dependent manner (Mattock et al., 2001b). In S. pombe, the effects of EGFP-pep10 and EGFP-mut10 were indistinguishable, indicating that the effect of these peptides is dependent upon the cyclin-binding motif rather than their PCNA interaction. The cell cycle block resulting from the expression of these EGFP-peptide fusions was also different in character to that caused by the other PCNA-binding peptides, showing a phenotype suggestive of a block very early in mitosis. No cut phenotypes or other mitotic defects could be detected by microscopic examination of pEGFP-pep10 or pEGFP-mut10 transformants (data not shown). The observation that expression of EGFP-pep10 did not lead to a PCNA-dependent cell cycle block was surprising, as expression of full length human p21(WAF1/Cip1) in S. pombe does result in a PCNA-dependent cell cycle block (Tournier et al., 1996). It is possible that this reflects the ability of full-length protein will be capable of binding to multiple protein targets. The pep10 sequence when fused to EGFP may act as a more “promiscuous” CDK-cyclin inhibitor than when the same sequence is expressed as part of the full-length p21(WAF1/Cip1) protein. Comparing the EGFP-pep10/EGFP-mut10 effects in human and S. pombe cells, these results may suggest that in human cells, competitive inhibition of PCNA function may be the rate-limiting step in regulating the cell cycle, compared to inhibition of cyclin function through the cyclin binding motif. The results shown here suggest that in S. pombe the converse is true.
We have shown using PCNA as a model protein that it is possible to use two-hybrid screening to identify peptides which bind to physiologically relevant protein interaction domains. We have identified a peptide which binds to PCNA from a range of species which shows functional activities in both a species of interest (human cells) and a model system (S. pombe). We have compared the activities of peptides derived from protein sequences (pep10), screening (pep102) and rational design (con1) in both human cells and in S. pombe. We find that, in terms of functional activity, the rationally designed peptide performed best in functional assays in both organisms. It is probable that screening of a more complex library would yield higher affinity peptides. Peptide stability was an important issue: short sequences when fused to a scaffold protein could result in widely differing steady state levels. Analysis of the peptide derived from human protein sequence was complicated by secondary binding activities. In this case, it was possible to delineate the two activities as the residues involved in each have been extensively characterised. Non-protein derived peptide sequences can, therefore, be more tractable as functional aptamers, as they do not contain secondary functional motifs. These results validate the use of both rational design and forward genetic screening for transdominant inhibitors of the cell cycle in yeast systems as potential leads for therapeutic compounds.
Materials and Methods
Plasmid expression constructs and yeast two-hybrid methods
Manipulations of E.coli and DNA and the growth and maintenance of S.cerevisiae were performed according to standard methods (Ausubel, 1997). Two-hybrid screening and analysis was carried out as previously described (Warbrick et al., 1995). Units of β-galactosidase were calculated as: Units = (1000 × OD420)/(t × v × OD600) where t = time of reaction in minutes and v = volume of culture in ml.
Peptide analysis
Twenty amino acid peptides were linked via residues SGSG at the amino terminus to biotin (Chiron Mimotopes, Australia).
pep10 KRRQTSMTDFYHSKRRLIFS
mut10 KRRQTSATDAYHSKRRLIFS
pogo KLFNLHINSAVLQKKITDYF
pep102 DPDCGCSRRGCVLRMIRRRN
pep103 DPLWCQSAVFLIRMWLRSRN
pep102j DPSRCRMGCDRGRLIRCVRN
pep103j DPMVWRILQASLCFSRWLRN
pep102s DCGCSRRGCVLRMIRR
pep103s LWCQSAVFLIRMWLRS
Con1 SAVLQKKITDYFHPKK
unrelated PESVELKWSEPNEEELIKFM
ELISA analysis of PCNA binding to peptides and peptide affinity capture experiments were carried out as previously described (Warbrick et al., 1995). In peptide competition experiments, non-biotinylated (free) peptides were added to the diluted cell extracts before incubation with the immobilised, biotinylated peptides. The unrelated control peptide used for competition was KPVRLPSIQAIPCAP. The final concentrations of competing peptides were 1000μg/ml, 100μg/ml, 10μg/ml, 1μg/ml and 0.1μg/ml.
Expression of EGFP constructs in human cells
pEGFP-pep102 and pEGFP-con1 were constructed from pEGFP-C3 (Clonetech) to express sequences MDCGCSRRGCVLRMIRR and SAVLQKKITDYFHPKK, respectively. pEGFP-pep10 and pEGFP-mut10 were as previously described (Mattock et al., 2001b). Cells were grown at 37°C and 5% carbon dioxide in DMEM (Dulbecco’s Modified Eagle’s Medium). Transfection was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were harvested following 24 hours post-transfection. Cells lysates were prepared as described (Mattock et al., 2001b). For immunoprecipitation experiments, equal quantities of lysate were pre-cleared with protein G-coupled agarose beads. The supernatants were incubated overnight at 4°C with 1μl of anti-EGFP rabbit antiserum Ab290 (AbCam). The immune complexes were captured with protein G-coupled agarose beads which were then washed extensively in PBS containing 0.2% (v/v) Tween-20. Samples were resuspended in 2x SDS-PAGE loading buffer, boiled and subjected to SDS-PAGE and Western blot analysis.
For clonogenic assays, 6 well plates were seeded with 2 - 5 × 104 cells per well and incubated overnight prior to transfecting with 6μg per well of plasmid DNA. The medium was replaced with fresh 24 hours post-transfection and G418 added to a final concentration of 1.0mg/ml at 48 hours post-transfection. Medium containing G418 was replaced as necessary. Cells were washed twice in PBS, fixed for 15 minutes in ice-cold methanol, then stained with 10% (w/v) Giemsa for 15 minutes and rinsed in water.
Expression of EGFP constructs in S. pombe
Standard methods were used for growth and manipulation of S. pombe (Moreno et al., 1991). All growth was at 30°C. The plasmid pREP1-pcn1+ is as previously described (Waseem et al., 1992). The plasmid pREP42-EGFPC-pep102 expresses the pep102 sequence at the C terminus of EGFP under the control of the nmt42 promoter and has ura4+ as a selectable marker for S. pombe (Craven et al., 1998). Plasmids were linearised with EcoRV to direct integration to the ura4 locus.
To make the vector series p4xG, p42xG and p82xG, the EGFP encoding sequence from pREP42-EGFPC was amplified with the primers GFPVecF1 and GFPVecR1. This sequence was then re amplified with the primers GFPVecF1 and GFPVecR2. The EGFP cassette was cloned as a XhoI – BamHI fragment into pREP4x, pREP42x and pREP82x to create p4xG, p42xG and p82xG respectively. p82xG gives an increased level of EGFP expression compared to pREP42-EGFPC.
GFPVecF1: GGGGCTCGAGTTAAATCATGGTGAGTAAAGGAGAAGAAC
GFPVecR1: CTCACTCACTCAGTCGACGCGGCCGCGGTACCTTTGTATAGTTCATCCATGCCATG
GFPVecR2: GGGGATCCATGCATGTTTAAATCTACTCACTCAGTCGACGCG
The pep10, mut10 and consensus I peptide (con1) encoding sequences were cloned from the corresponding pEGFP constructs described above as XhoI – BamHI fragments into SalI – BamHI digested p4xG, p42xG and p82xG. The pep102 sequence was cloned into these vectors from pEGFP-pep102 as a XhoI – BamHI fragment.
For the analysis of protein expression, log phase cell cultures were washed with sterile water and then inoculated into EMM minus thiamine and diluted as necessary into fresh medium. Viability was assessed by plating known numbers of cells onto YE medium and counting colony formation. To assess UV sensitivity, cells were plated in duplicate and one plate immediately exposed to 10J/m2 at 254nm using a CL-1000 UV Crosslinker (UVP). Cell lysates were prepared using Y-PER yeast protein extraction reagent (Pierce) according to the manufacturers instructions. For microscopy, dried films of cells were mounted in Hydromount (National Diagnostics) containing DAPI at 1μg/ml. Slides were examined with a Zeiss Axiovert fluorescence microscope, with image analysis using AxioVision software. For FACS analysis, cells were fixed and stained with propidium iodide (Sazer & Sherwood, 1990). Cells were analysed using a Becton Dickinson FACScan.
Acknowledgements
I would like to thank Tony Carr for the nmt-EGFP plasmid and Daniella Zheleva for purified human PCNA. I would also like to thank Sylvie Tournier for plasmids and helpful discussions and Alison Sparks for her help with FACS analysis. I would like to thank all my colleagues for their help and support, especially David Lane and the members of his laboratory. This work was supported by the Association for International Cancer Research and the Cancer Research Campaign.
REFERENCES
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG., Jr. Mol Cell Biol. 1996;16:6623–33. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Mol Biol Cell. 1994;5:147–60. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Current Protocols in Molecular Biology. Wiley; [Google Scholar]
- Cayrol C, Knibiehler M, Ducommun B. Oncogene. 1998;16:311–20. doi: 10.1038/sj.onc.1201543. [DOI] [PubMed] [Google Scholar]
- Chen IT, Smith ML, O’Connor PM, Fornace AJ., Jr. Oncogene. 1995;11:1931–7. [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJ, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H, Kelman Z, Dean FB, Pan ZQ, Harper JW, Elledge SJ, O’Donnell M, Hurwitz J. Proc Natl Acad Sci U S A. 1994;91:8655–9. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Hall PA, Kearsey JM, Coates PJ, Norman DG, Warbrick E, Cox LS. Oncogene. 1995;10:2427–33. [PubMed] [Google Scholar]
- Jonsson ZO, Hubscher U. Bioessays. 1997;19:967–75. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- Kamb A, Caponigro G. Curr Opin Chem Biol. 2001;5:74–7. doi: 10.1016/s1367-5931(00)00160-5. [DOI] [PubMed] [Google Scholar]
- Kelman Z. Oncogene. 1997;14:629–40. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Hurwitz J. Trends Biochem Sci. 1998;23:236–8. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- Kontopidis G, Wu SY, Zheleva DI, Taylor P, McInnes C, Lane DP, Fischer PM, Walkinshaw MD. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0406540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Nurse P. Nature. 1987;327:31–5. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Dowbenko D, Lasky LA. J Biol Chem. 2002;277:11352–61. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- MacNeill SA, Moreno S, Reynolds N, Nurse P, Fantes PA. Embo J. 1996;15:4613–28. [PMC free article] [PubMed] [Google Scholar]
- Mattock H, Jares P, Zheleva DI, Lane DP, Warbrick E, Blow JJ. Exp Cell Res. 2001a;265:242–51. doi: 10.1006/excr.2001.5181. [DOI] [PubMed] [Google Scholar]
- Mattock H, Lane DP, Warbrick E. Exp Cell Res. 2001b;265:234–41. doi: 10.1006/excr.2001.5160. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. Mol Cell Biol. 2000;20:1206–18. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Embo J. 1999;18:3834–44. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Warbrick E, Fantes PA, MacNeill SA. Embo J. 2000;19:1108–18. doi: 10.1093/emboj/19.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S. Mol Cell Biol. 2001;21:5644–57. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Sherwood SW. J Cell Sci. 1990;97(Pt 3):509–16. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O’Connor PM, Fornace AJ., Jr. Science. 1994;266:1376–80. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- Tournier S, Leroy D, Goubin F, Ducommun B, Hyams JS. Mol Biol Cell. 1996;7:651–62. doi: 10.1091/mbc.7.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T. Front Biosci. 1999;4:D849–58. doi: 10.2741/tsurimoto. [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B. Nature. 1994;369:574–8. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Warbrick E. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Heatherington W, Lane DP, Glover DM. Nucleic Acids Res. 1998;26:3925–32. doi: 10.1093/nar/26.17.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. Curr Biol. 1995;5:275–82. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- Waseem NH, Labib K, Nurse P, Lane DP. Embo J. 1992;11:5111–20. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhang P, Liu L, Lee MY. Biochemistry. 2001a;40:4512–20. doi: 10.1021/bi010103+. [DOI] [PubMed] [Google Scholar]
- Xu X, Leo C, Jang Y, Chan E, Padilla D, Huang BC, Lin T, Gururaja T, Hitoshi Y, Lorens JB, Anderson DC, Sikic B, Luo Y, Payan DG, Nolan GP. Nat Genet. 2001b;27:23–9. doi: 10.1038/83717. [DOI] [PubMed] [Google Scholar]
- Yang M, Wu Z, Fields S. Nucleic Acids Res. 1995;23:1152–6. doi: 10.1093/nar/23.7.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheleva DI, McInnes C, Gavine AL, Zhelev NZ, Fischer PM, Lane DP. J Pept Res. 2002;60:257–70. doi: 10.1034/j.1399-3011.2002.21014.x. [DOI] [PubMed] [Google Scholar]
- Zheleva DI, Zhelev NZ, Fischer PM, Duff SV, Warbrick E, Blake DG, Lane DP. Biochemistry. 2000;39:7388–97. doi: 10.1021/bi992498r. [DOI] [PubMed] [Google Scholar]