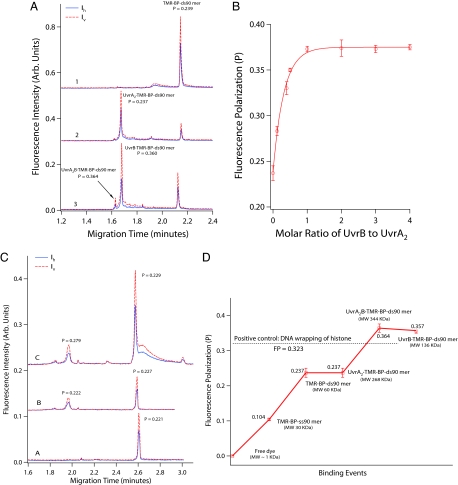
Fig. 1.
CE-LIFP analysis of the interaction of fluorescent DNA substrates with E. coli UvrA and B proteins. (A) CE-LIFP analysis of TMR-BP-ds90mer and its binding to UvrAB, showing a large increase of fluorescence polarization (P). The reaction solutions contained 10 nM TMR-BP-ds90mer (trace 1); 10 nM TMR-BP-ds90mer and 20 nM UvrA (trace 2); or 10 nM TMR-BP-ds90mer, 20 nM UvrA, and 40 nM UvrB (trace 3). The solid and dashed lines represent horizontally (Ih) and vertically (Iv) polarized fluorescence, respectively. (B) UvrB concentration-dependent increase of fluorescence polarization of UvrAB-TMR-BP-ds90mer complex. TMR-BP-ds90mer and UvrA were kept at 10 and 20 nM, respectively, and UvrB was varied from 0 to 40 nM. (C) CE-LIFP analysis of the mixtures of unmodified TMR-ds90mer with UvrA and UvrB proteins, showing a lack of increase in fluorescence polarization after UvrB binding to undamaged DNA. Shown are 10 nM TMR-ds90mer (trace A), 10 nM TMR-ds90mer + 20 nM UvrA (trace B), and 10 nM TMR-ds90mer + 20 nM UvrA + 40 nM UvrB (trace C). (D) Fluorescence-polarization values of DNA–protein intermediates generated along the initial steps of the E. coli NER pathway from TMR-BP-ds90mer, UvrA binding to TMR-BP-ds90mer, UvrA and UvrB binding to TMR-BP-ds90mer, and UvrB binding to TMR-BP-ds90mer. Fluorescence-polarization values for free dye (TMR) and TMR-BP-ss90mer were also measured for comparison.