Abstract
The major structural components of HIV are synthesized as a 55-kDa polyprotein, Gag. Particle formation is driven by the self-assembly of Gag into a curved hexameric lattice, the structure of which is poorly understood. We used cryoelectron tomography and contrast-transfer-function corrected subtomogram averaging to study the structure of the assembled immature Gag lattice to ≈17-Å resolution. Gag is arranged in the immature virus as a single, continuous, but incomplete hexameric lattice whose curvature is mediated without a requirement for pentameric defects. The resolution of the structure allows positioning of individual protein domains. High-resolution crystal structures were fitted into the reconstruction to locate protein–protein interfaces involved in Gag assembly, and to identify the structural transformations associated with virus maturation. The results of this study suggest a concept for the formation of nonsymmetrical enveloped viruses of variable sizes.
Keywords: cryoelectron tomography, virus assembly, contrast transfer function, capsid, retrovirus
HIV assembly is driven by the 55-kDa Gag polyprotein which forms a curved protein lattice at the plasma membrane (1, 2). Particle release also requires recruitment of components of the cellular endosomal sorting complex required for transport (ESCRT) machinery to the budding site (1, 2). The virus is initially produced in an immature form, where the Gag polyproteins form a radially arranged layer underneath the viral membrane. The N-terminal matrix (MA) domain of Gag interacts with the membrane and the C-terminal nucleocapsid (NC) and p6 domains are located toward the center of the particle (3). Between MA and NC, the capsid (CA) domain arranges into a regular lattice, the dimensions of which are conserved between different retroviruses (4). Activation of the viral protease (PR) leads to cleavage of Gag and dramatic morphological rearrangements, termed maturation, which are required for infectivity (5). After maturation, the ribonucleoprotein condenses in the center of the particle, where it is surrounded by a characteristic cone-shaped core shell formed from CA. MA remains associated with the membrane. The structure of the CA lattice in the mature cone has been studied through cryoelectron microscopy (cEM) and tomography (cET) of mature-like in-vitro-assembled particles (6) and mature virions (7–9). Most recently, electron crystallography revealed the structure to a high resolution at which alpha helices could be resolved (10). The mature lattice formed from hexamers of the N-terminal CA (N-CA) domain, linked by C-terminal CA (C-CA) dimers at approximately the same radial position within the core. The N-CA hexamers have a small central hole, and the lattice is stabilized by a defined N-CA–C-CA interaction interface.
In contrast to the detailed understanding of the mature lattice, there is no higher-resolution reconstruction of the immature Gag lattice. By using cEM, the CA domain in immature HIV has been found to display a curved 2D lattice with an inter-hexamer distance of 8.0 nm (11). The lattice exhibits local hexagonal, but lacks global icosahedral, order (12). Fuller et al. (12) suggested that small patches of locally ordered Gag form facets on the virus surface. Recently, Wright et al. (13) studied the structure of immature HIV by cET. They reported the Gag lattice to be incomplete, adopting a patchwork arrangement. A number of possible models could explain this arrangement and intermediates are also possible. At one extreme, Gag could form a closed spherical lattice through the regular inclusion of 12 pentameric defects, the strict structure of which would be lost after budding. At another extreme, “islands” of Gag hexamers could gather at the membrane and become enclosed in the budding virion. By using a combination of electron tomography (ET) and scanning-transmission EM, we recently also found the Gag lattice to be incomplete in the immature virus. In our study, the lattice appeared to form a continuous shell underlying ≈60% of the spherical membrane of the virus (14). Mass measurements revealed that the regions without a regular lattice were not due to a disordered Gag lattice, but to lack of Gag.
Features that are present in multiple copies in reconstructions from cET, such as envelope spikes or the unit-cell of the viral protein lattice, can be extracted, aligned, and averaged together in a procedure called subtomogram averaging. The resulting average can be interpreted at a higher resolution than the original tomogram. Higher-resolution reconstructions have been produced of glycoprotein spikes by using subtomogram averaging (15–17). These reconstructions have allowed positioning of protein domains within the density. It is necessary, however, to take into account the contrast-transfer-function (CTF) of the EM to obtain high-resolution structures from cEM data (18). CTF correction has not yet been demonstrated for subtomogram averaging techniques. This limits the interpretable data to a resolution of ≈2 nm in the best published reconstructions.
Despite several recent advances, many aspects of immature HIV assembly and structure remain unclear. It is not known whether the immature Gag shell is formed by a single hexameric lattice or by multiple patches, nor how the assembling hexagonal lattice forms into a curved structure. Furthermore, it is not clear how Gag domains are packed together in the immature lattice, and which protein-interaction interfaces drive assembly. Here we have used cET and CTF-corrected subtomogram averaging to study the structure of immature HIV and in-vitro-assembled immature-like particles. Mapping the positions of hexameric unit cells in these particles revealed the presence of a single continuous, but incomplete, hexameric lattice, closed by the incorporation of irregular defects. The resolution of the reconstruction of the immature Gag lattice is sufficient to allow us to position individual CA domains within the lattice.
Results
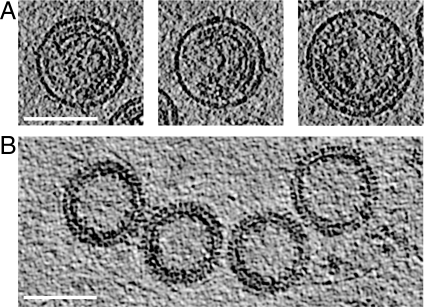
Highly purified immature HIV-1 particles were imaged by cET at a range of different defocuses, as described in SI Text, and reconstructed in 3 dimensions (Fig. 1A). The virus particles were approximately spherical with variable diameters, consistent with earlier observations (3, 11). All virions analyzed showed an incomplete Gag lattice underneath the plasma membrane, as previously reported (13, 14). As a complementary approach, immature-like particles lacking a membrane were produced by in vitro assembly of a bacterially expressed Gag protein, missing most of the MA and the entire p6 domain, and analyzed in parallel. These particles, which are known to exhibit the same protein lattice as immature HIV-1 (11), were substantially more complete than immature viruses produced in tissue culture (Fig. 1B).
Fig. 1.
Sections through tomograms of (A) immature virus particles and (B) in-vitro-assembled Gag particles. Scale bar, 100 nm.
To obtain detailed information about the local and global structure of the Gag lattice, subtomograms containing small regions of the membrane and underlying density were extracted from the tomograms and subjected to alignment and averaging procedures. These procedures provide 2 types of output describing the Gag lattice structure. Firstly, the final averaged reconstruction reveals the local structure of the Gag lattice at a resolution sufficient to visualize individual protein domains. Secondly, the final positions of the subtomograms after translation and rotation reveal the global arrangement of Gag. Comparison of the global arrangement and the local structure can be used to verify the success of the alignment procedures.
Global Arrangement of the Lattice.
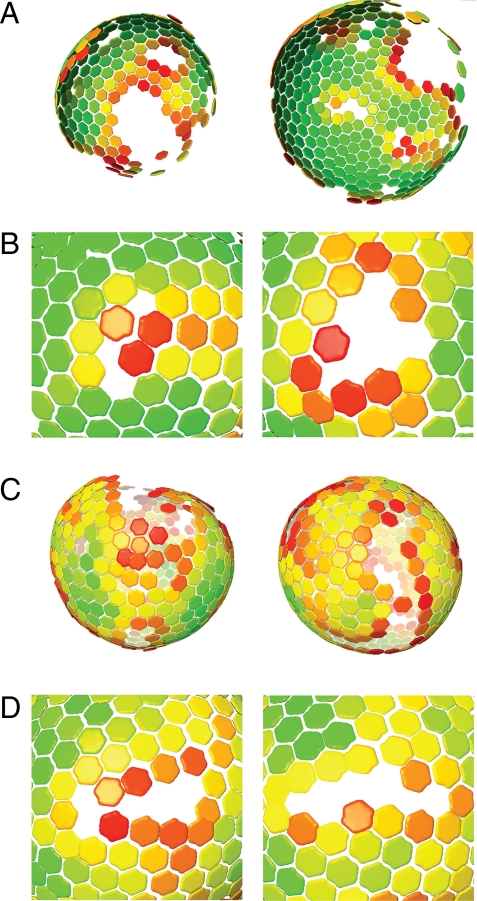
Global lattice maps of immature HIV-1 particles purified from the medium of cultured cells were created by placing hexamers at the final positions and orientations to which the alignment converged (Fig. 2A). This provided a visual representation of the positions of the Gag hexamers within a particular virus particle. All particles observed were substantially incomplete, with large parts of the viral surface lacking an ordered arrangement of Gag. Inspection of the large, disordered regions in the original tomograms showed no significant protein density underneath the membrane, confirming that this disorder results from the absence of Gag, and not from the presence of disordered Gag.
Fig. 2.
Global lattice maps of HIV particles. Positions of hexameric unit cells are marked with hexamers. Hexamers are colored according to cross-correlation on a scale from low (red) to high (green). Maps are shown in perspective such that hexamers on the rear surface of the particle appear smaller. (A) Lattice maps for immature HIV particles. The side of the particle toward the viewer lacks ordered Gag. (B) Close-up of defects in immature Gag lattice. (C) Lattice maps for in-vitro-assembled Gag particles. (D) Close-up of defects in in-vitro-assembled particle lattice.
A uniform, extended, hexameric lattice is flat. Curvature of a hexameric lattice can be achieved by the insertion of pentameric defects, as is the case in the mature viral core, or by breaking the uniformity of the lattice in other ways. Inspection of the ordered regions in the immature virus particles showed that the area of ordered Gag was made up of a single, interconnected hexameric lattice in each virus particle. No agglomeration of small islands of Gag protein was observed. Curvature in the hexameric lattice was mediated by the incorporation of defects into the lattice. The distribution of hexamers around the defects did not suggest any preference for pentameric defects. Instead, defects adopted a number of different geometries, as illustrated in Fig. 2B.
In-vitro-assembled Gag particles were subjected to the same imaging and analysis procedures as the immature virus. Global lattice maps of the in-vitro-assembled particles confirmed them to be considerably more complete than the virus (Fig. 2C). Curvature was again mediated by the incorporation of a range of differently shaped defects (Fig. 2D) rather than by the exclusive incorporation of pentameric defects.
Local Structure of the Lattice.
The lipid bilayer in HIV-1 particles appears different in areas where Gag is bound underneath, compared with areas where no Gag is bound (Fig. 1A), which has been suggested to represent a difference in the thickness of the bilayer (13). To compare the membrane in the presence or absence of Gag, the subtomograms extracted from the tomograms of immature virus particles were segregated into 2 classes according to the presence or absence of Gag (see SI Text). These 2 classes were separately averaged (Fig. 3). The inner leaflet of the bilayer appeared thicker in regions where a Gag lattice is present (Fig. 3 Left) than in regions where no Gag is bound to the membrane (Fig. 3 Right). This thickening is likely due to the presence of the membrane-associated MA domain (Fig. 3, compare Left and Right). No obvious difference in the separation of the inner and outer leaflets was observed between the 2 reconstructions. The measured separation of the leaflets in bilayer regions lacking Gag was 4 nm. The apparent change in bilayer thickness seen in Fig. 1A and in other published images (13) likely results from a combination of the CTF of the microscope and low pass filters applied to the images. These effects are illustrated in a simulation presented as SI Text and Fig. S1. Fig. S1 shows that the presence of a protein layer apposed to a membrane leads to an apparent increase in bilayer thickness at further-from-focus conditions.
Fig. 3.
Part of central section through reconstruction of immature virus particle from regions with Gag bound to the membrane (Left) or where no Gag is bound to the membrane (Right). Density is dark on a light background. The inner leaflet of the bilayer is thicker in regions where Gag is present, but the bilayer spacing remains constant.
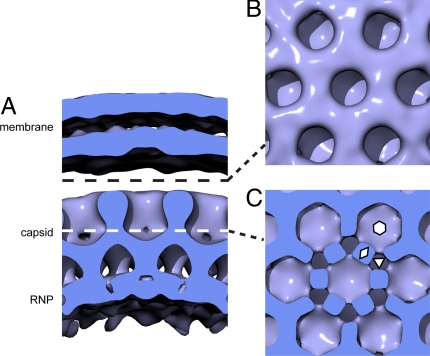
The reconstruction of the local Gag lattice revealed a hexameric unit cell with a spacing of 8 nm at the C-CA domain (Fig. 4 and Fig. S2A). The radial positions of the protein domains were assigned based on Wilk et al. (3). At the expected radial position of N-CA, the density forms hexameric rings with large central holes, as previously described in Wright et al. (13) (Fig. 4B). At the expected radius of C-CA, well defined 2-fold densities were seen, which we interpret as corresponding to dimers of C-CA (Fig. 4C). At this radius there are holes at the local 3-fold symmetry axes. The 2-fold densities link together at the local 6-fold symmetry axes in the reconstruction to form pillar-like densities that descend into the RNP layer. This density is consistent with the proposed 6-helix bundle in the spacer peptide 1-NC linker region (13, 19–) but at the current resolution could also accommodate other arrangements. The gap between N-CA and the MA-layer and the holes in the N-CA layer are both sufficiently large enough to accommodate the cellular protein cyclophilin A, which is incorporated into HIV-1 by binding to N-CA at a ratio of 1:10 compared with Gag (22, 23).
Fig. 4.
Surface rendering of reconstruction of immature virus particle. (A) Surface cut perpendicular to the membrane to reveal the 2 membrane leaflets, the CA layer, and the RNP. (B) Surface cut tangential to the membrane at a radius indicated by the dashed black line in A, and looking down on the N-CA domains. (C) Surface cut tangentially along the white dashed line in A, through the C-CA domains. Hexagon, triangle, and rhombus indicate examples of 6-fold, 3-fold, and 2-fold symmetry axes, respectively.
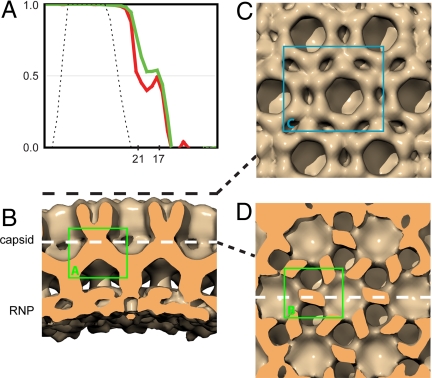
The local Gag lattice was also reconstructed from tomograms of in-vitro-assembled particles. The absence of the membrane, the higher completeness of the particles, and the absence of other viral and cellular components provided the opportunity to obtain a higher-resolution reconstruction from these particles. Thirteen particles from the 3 of the best tomograms were combined after CTF correction to produce a reconstruction with a resolution in the central C-CA region of ≈17 Å (Fig. 5A, see also Figs. S2B and S3). This resolution is higher than previously obtained by using subtomogram averaging techniques. The reconstruction showed clear features in both the N- and C-terminal domains (Fig. 5 B–D), which could be assigned to individual protein domains. The unit-cell size and the low resolution of the Gag lattice in the in-vitro-assembled particle are the same as those in the immature particle (compare Fig. 4A and Fig. 5B), confirming our previously published observation that the in-vitro-assembled and immature Gag lattices share the same structure (11).
Fig. 5.
Reconstruction of in-vitro-assembled Gag particles. (A) Fourier shell correlation plot, before (red) and after (green) CTF correction. The black dotted line indicates the band-pass filter applied during the reconstruction process. The resolutions measured according to the 0.5 criterion are 21 Å before CTF correction, and 17 Å after CTF correction. (B) Surface rendering sectioned perpendicular to the particle surface. (C) Surface cut tangential to the surface at a radius indicated by the dashed black line in B, looking down on the N-CA domains. (D) Surface cut tangentially along the white dashed line in B, through the C-CA domains. Colored boxes refer to the regions illustrated in Fig. 6 A–C.
For both the immature virus particles and the in-vitro-assembled Gag particles, the presence of large areas of hexagonal order in the global lattice maps verifies that the alignment procedures used to produce the local lattice reconstructions have been successful, and that the final reconstruction does not result from noise alignment.
Fitting Atomic Resolution Structures Into the Density.
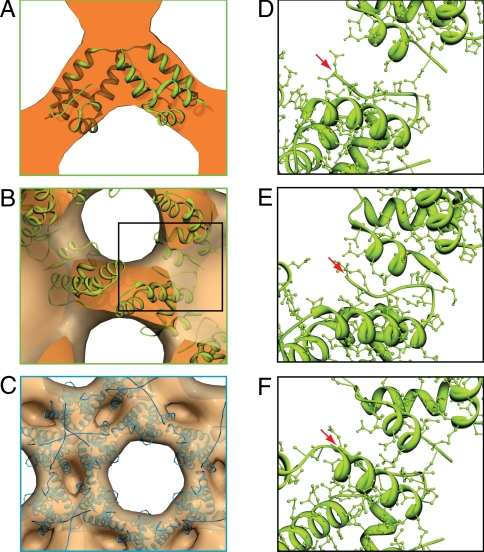
We next interpreted the reconstruction by fitting atomic resolution structures of individual protein domains into the higher-resolution in vitro structure (Fig. 6). Four different structures have been published for the HIV C-CA dimer and they were individually fitted into the density and compared. Dimer 1 [PDB3ds5 (24), equivalent to PDB 1a43 (25)] and dimer 2 [PDB1a8o (25)] possess the same basic monomer structure and dimer interface, but exhibit different crossing angles of the dimerising helices. Dimer 3 (PDB3ds2) represents the structure of the C-CA dimer in complex with the capsid assembly inhibitor (CAI) (24, 26). This dimer structure is also adopted by C-CA variants carrying mutations in the CAI-binding pocket. Such mutations do not affect immature virus formation, although they do abolish HIV-1 infectivity. Dimer 4 (PDB2ont) corresponds to the structure of the domain-swapped C-CA dimer described by Ivanov et al. (27), which has been suggested to be relevant for the immature lattice. All dimers were fitted into the density by using an automated procedure while constraining the 2-fold symmetry axis of the dimer to be coincident with the local 2-fold axes in the structures. During fitting, all local symmetry axes in the density were applied to allow monitoring of molecular clashes.
Fig. 6.
Fitting crystal structures into the in vitro particle reconstruction. (A) The best identified fit of the C-CA dimer PDB3ds2 into the region boxed in Fig. 5B. (B) The same fit viewed as boxed in Fig. 5D. (C) One possible fit of the N-CA domain, viewed as boxed in Fig. 5C. (D and E) Close-ups of the inter-dimer contacts highlighted with the black box in 6B. (D–F) The fits shown are for Dimer 2, PDB1a8o (D), Dimer 3, PDB3ds2 (E), and Dimer 4, PDB2ont (F). Red arrows indicate the position of the N-terminal residue of C-CA.
The quality of the 4 fits was compared using 3 criteria: The maximum cross-correlation value between the fitted structure and the density; the number of molecular clashes made between the structure and its neighbors in the assembled lattice; and the radial distance between aspartate 152 of the C-CA domain and serine 146 of the N-CA domain, which must be consistent with the 2 domains being part of 1 polypeptide chain. Table 1 and Fig. 6 D–F summarize the comparison between the fits of the 4 crystal structures. The cross-correlation values are similar for all 4 dimers. The number of contacts between neighboring dimers around the hexamer was particularly informative. The precise conformation observed for the exposed side-chains in a crystal structure is unlikely to represent the side-chain conformations which would be formed at a protein–protein interface. We therefore assume that 2 domains fitted in sufficiently close enough proximity to interact should exhibit multiple side-chain contacts and clashes. Fitting the structure of either dimer 1 or 2 gave few or no contacts between neighboring dimers around the hexamer (Fig. 6D). It is therefore unlikely that they would be able to form meaningful interdimer interactions. In contrast, dimer 3 (Fig. 6E) and to a lesser extent dimer 4 (Fig. 6F) showed multiple contacts across this interface, consistent with interdimer contacts around the hexamer.
Table 1.
Statistics for fitting of C-terminal CA dimers into the reconstruction
| PDB code | Cross-correlation | Number of contacts | Radial distance, Å |
|---|---|---|---|
| PDB3ds5 | 0.56 | 8 | 21 |
| PDB1a8o | 0.59 | 0 | 18 |
| PDB3ds2 | 0.59 | 118 | 19 |
| PDB2ont | 0.62 | 23 | 23 |
Number of contacts is number of atomic contacts between neighboring dimers around the hexamer. Radial distance is the difference in radial position between S146 in the N-terminal domain and D152 in the C-terminal domain. If these were linked by a beta-strand, they would be separated by 21 Å.
Dimers 1 and 4 position the N-terminal residues of C-CA relatively far from the density corresponding to N-CA. When viewed from N-CA, down the 2-fold axis, the N-terminal residues of C-CA in these 2 dimers sit below helix 1 and the helix 1-2 loop (red arrow in Fig. 6F). The larger distance and this occlusion make it difficult to satisfactorily position the N-CA within the density in a position where it is close enough to be joined to C-CA. In contrast, dimers 2 and 3 position the N-terminal residues of C-CA to the side of the protein (Fig. 6 D and E) where they are better positioned to link to N-CA.
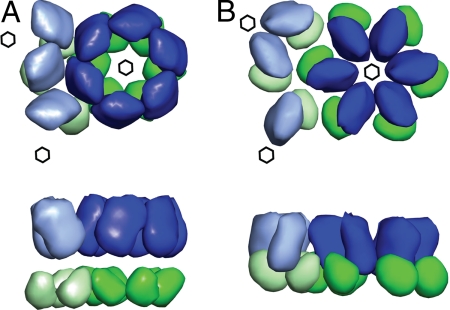
The fitting of the N-CA structure is complicated by the presence of amino acids in the assembled protein lattice, which were not ordered in the high resolution structure of the protein. These residues may or may not contribute significantly to the lattice reconstruction, depending on whether they are well-ordered in the lattice. PDB1l6n (28), an NMR structure of the N-CA domain, together with MA, was edited to remove the attached MA domain and manually fitted into the density. Only fits which placed the C-terminal end of the protein toward the C-CA domain, and which avoided overlap between neighboring domains in the lattice, were considered. Two general orientations of the molecule were most consistent with the density. The N-CA domain has an “arrowhead-like” shape. The 2 orientations direct the point of the arrowhead toward the center of the particle, and one of the flat faces of the arrowhead toward the six-fold axis, with some flexibility around this position. One of these general orientations is illustrated in Fig. 6C, the other is an ≈180 ° rotation of the domain in the plane of the image. The density is tilted slightly such that the hole at the 6-fold is smaller at the outside of the particle than at the lower radii. The proposed high-resolution models for the hexameric arrangements of the N-CA domain place one of the sharp edges of the arrowhead toward the six-fold axis (29, 30), similar to its positioning in the mature lattice (Fig. 7B). These arrangements are not consistent with our immature structure, where one of the flat sides of the arrowhead is oriented toward the 6-fold axis (Fig. 7A). Taken together, these results suggest substantial differences in the arrangement of both N-CA and C-CA between the immature and mature CA lattices.
Fig. 7.
Schematic of the structural changes taking place during maturation. (A) The immature arrangement of the N-terminal (blue) and C-terminal (green) domains of CA, viewed from outside the particle (Top), and rotated 90 ° around the horizontal axis (Bottom). Domains from neighboring hexamers are indicated in lighter colors. Six-fold lattice positions are marked by hexagons. (B) The mature arrangement, with domains positioned according to ref. 10.
Discussion
The Assembly Process.
For reasons of genetic economy, viruses assemble their protein coats from multiple copies of a small number of distinct proteins. The assembly of multiple copies of identical proteins leads to particles with local (e.g., hexagonal) and often global (e.g., helical or icosahedral) symmetry. The formation of a closed shell from a protein which forms a hexagonal lattice is most elegantly achieved by the incorporation of 12 evenly spaced pentamers or pentameric defects, yielding a particle with icosahedral symmetry. Such an arrangement can be adopted by in-vitro-assembled Rous Sarcoma virus CA proteins (31). The recently published high-resolution EM structure of this particle (31) provides strong support for the presence of CA pentamers in mature retroviral cores. In mature cores, uneven spacing of the pentamers within the hexameric lattice can give rise to a range of different core geometries (32). In contrast to mature cores, pentamers have not been observed in the immature Gag lattice, but have generally been assumed to be present to mediate curvature of the lattice.
Immature retroviruses are known to possess an unusual degree of irregularity. Gag proteins form a hexagonal lattice that gives rise to a particle lacking icosahedral symmetry. This lattice is also substantially incomplete. The global lattice maps presented here allow us to draw 2 further conclusions about the assembly process, suggesting a model to explain lattice curvature. Firstly, the hexameric lattice within the immature virus particles as well as the in-vitro-assembled Gag particles is continuous. It does not consist of a patchwork arrangement of smaller areas of hexagonal order. Rather, a single hexameric lattice can be traced in the majority of Gag particles. Secondly, the defects which are inserted into the hexameric lattice to permit curvature are not uniform in their shape. The curvature of the lattice is not mediated by the irregular incorporation of pentameric defects, but rather by the irregular incorporation of irregular defects.
These observations suggest a simple model for HIV-1 particle formation, in which Gag assembles into a hexameric lattice that grows with an inherent curvature and that incorporates new protein molecules stochastically. One attractive, simple model for inherent curvature would be that the N-CA domains pack to form a hexameric lattice with a preferred unit-cell size, and the adjoining C-CA domains pack to form a hexameric lattice with a slightly smaller preferred unit-cell size. As any curved hexameric lattice grows, the hexamers further from the center of the growth point become increasingly tightly packed. This can be envisaged by wrapping a flat hexameric lattice over the surface of a sphere, as illustrated in Fig. S4. This mechanism will naturally lead to the incorporation of defects, because a point will be reached during growth where it is more favorable to leave gaps in the growing assembly than to pack the protein at increasing density. The extent to which such a curved hexameric lattice would grow between defects, and therefore the size of the particle, is largely dictated by the inherent curvature and flexibility of the protein. In vitro, the lattice grows to an almost complete spherical shell, whereas in virus producing cells the ESCRT machinery is recruited to the growing bud and completes the budding process leaving a large gap in the spherical shell of the released immature virion (14). In contrast with the mature core, no specific proteins or Gag arrangements are required to define either pentameric defects or the size of the immature particle. This mechanism is one of the simplest that can be conceived for formation of spherical enveloped viruses of variable sizes, but to our knowledge it has not been shown for any other virus type.
Subtomogram Averaging.
Subtomogram averaging methods allow the structures of macromolecular complexes to be solved in situ. The resolution attainable depends on the flexibility of the structure, on the number of copies which can be averaged, and on the accuracy with which the individual subtomograms can be aligned. Wright et al. (13) used averaging methods to produce a reconstruction of the immature HIV-1 Gag lattice, revealing hexagonal rings at the radius of CA. Further toward the center of the particle, a second hexagonally arranged layer was seen with strong density regions directly below the holes in the CA lattice. At the resolution obtained, individual protein domains could not be resolved. Therefore, many possible arrangements of the N- and C-terminal domains of CA are consistent with these data. Our results are consistent with the conclusions by Wright et al. (13), but provide a higher resolution and can thus be interpreted by fitting high-resolution structures. Previously, reconstructions have been carried out without CTF correction. For good samples, the resolution is limited by the first node of the CTF. Data should be filtered to this resolution to prevent the incorporation of phase-inverted information. For a tomogram collected at a defocus of −2 μm on a 300-kV microscope, this node falls at ≈2 nm. Collecting data closer to focus where this node falls at higher resolution gives low signal-to-noise data, which is difficult to align. Correction of the CTF in an image requires measurement of the defocus of the image, which has proved difficult in cET, because the images are tilted and, more critically, because the signal-to-noise ratio of individual images is low. Here we have corrected the CTF of the data based on the measured mean defocus of the series and on Fourier comparison of reconstructions generated from different tomograms taken at different defocuses. This correction has allowed us to incorporate data past the first node of the CTF and produce a reconstruction at a resolution of 17 Å, thereby providing proof of principle for obtaining reconstructions of protein complexes in vivo at higher resolution in the future.
Gag–Gag Interactions During Assembly.
To aid interpretation of the density, we have fitted the high-resolution crystal structures of individual CA domains into the reconstruction. Four different crystal structures for the dimeric C-CA domain were considered. Dimers 1 and 2 show very little protein interface in the intrahexameric contacts. This situation is difficult to reconcile with the well-defined hexameric unit-cell size at this radius in the particle. Dimer 1 has been shown to be most similar to the C-CA structure in the mature assemblage (10), although none of the published structures gave a perfect fit in this case. Dimer 3 in particular, but also dimer 4, both allow some protein contacts around the hexamer. Although the swapped dimer (dimer 4) is a very attractive model and would also be consistent with experimental data, one has to keep in mind that having a swapped dimer in the immature lattice and an unswapped dimer (dimer 1) in the mature lattice would require the coordinated unfolding of a large interaction interface [≈1,620 Å2 (27)] upon virion maturation. The low-radius, occluded position of the N-terminal residues of C-CA in dimer 4 is also difficult to reconcile with a connection to N-CA. Dimer 3 is clearly incompatible with formation of the mature core, but mutations forcing C-CA to crystallize in this conformation had no influence on assembly of immature-like particles in vitro or on the formation and morphology of immature HIV in tissue culture. Although the current resolution of the structure does not allow us to unequivocally identify which of the dimer structures (if any) corresponds to the conformation that the C-CA domain adopts in the immature lattice, these arguments lead us to prefer dimer 3 as being closest to the immature structure. In this case, C-CA would undergo a conformation change upon maturation corresponding to the difference between dimer 3 and the mature form of the dimer. This change alters both the dimer interface and the CAI-binding pocket. Regardless of which dimer was considered, the best fit placed residues 153–159 (IRQGPKE) of one CA molecule close to residues 212–219 (EEMMTACQ) on the neighboring CA molecule in the hexamer, suggesting this region may be involved in interdimer interactions within the hexamer.
The N-CA domain is arranged around the 6-fold axis to give a large hole. The density is consistent with a model where one of the flat faces of the arrowhead-shaped N-CA domain faces the hole. The density is not consistent with the published high-resolution models for N-CA hexamers which place one of the sharp edges of the arrowhead toward the 6-fold axis (29, 30).
The positions of C-CA and N-CA in the immature CA lattice are therefore quite different from their positions in the mature lattice (compare Fig. 7 A and B). The N-CA domains are rotated and the hexamers are opened outward to give a large, central hole in the immature lattice. The C-CA dimers are in a separate layer toward the center of the particle with no clear N-CA–C-CA interaction interface. The C-CA dimers are also separated from one another to give a hole in the density at the 3-fold position of the lattice. The overall impression is that upon maturation, the N-CA domains rotate and move toward the 6-fold axis, which, combined with an increase in unit-cell size, creates a gap at the 2-fold positions. The C-CA domains compact to form a fatter dimer structure, which occupies this gap. Higher-resolution structures of the immature lattice will be required to unequivocally position atomic structures of individual Gag domains and precisely define contact sites. This goal should be reached through extension and optimization of the approaches described in this report. When combined with larger datasets, this will also permit detailed study of the structures of the edges and the defects in the assembling lattice.
Materials and Methods
Sample Preparation.
MT-4 cells were maintained in RPMI-1640, supplemented with 10% FCS and antibiotics. Infection with HIV-1 strain NL43 by coculture and purification of the virus was performed as described (33). Immature particles were obtained by adding 2 μM lopinavir 6 h after infection. Samples were inactivated by using 1% PFA for 1 h on ice as described (9). In-vitro-assembled HIV Gag particles were prepared as described (34, 35).
Initial Processing.
cET was performed as described in SI Text. Regions of tomograms containing individual viruses or in-vitro-assembled particles were extracted and subjected to further MATLAB-based (Mathworks) processing by using subtomogram averaging approaches developed from those described in Forster et al. (36). Fig. S5 gives a schematic overview of the subtomogram averaging method. From each virus or in-vitro-assembled particle, a set of overlapping subtomograms were extracted with the centers of the extracted subtomograms distributed evenly on the surface of a sphere. The sphere was defined to have a center corresponding to the particle center and a radius corresponding to the radial position of the CA layer of Gag. Initial starting models were generated by using the geometry of the sphere with no external reference, as described in SI Text.
Refinement and CTF Correction.
For immature virus, data from 3 virus particles were combined and run through further iterations of alignment and averaging to generate a final reconstruction. The in vitro particles were reconstructed to higher resolution by using higher-magnification tomograms. From each higher-magnification tomogram of in-vitro-assembled particles, 3–5 particles were extracted, and subtomograms were extracted from the particles. The subtomograms were iteratively aligned (in 6 dimensions, considering translation and rotation) and averaged, with the initial in-vitro-assembled particle model as a starting point. To avoid the possibility of noise alignment, a low-pass filter was applied during the alignment at a lower resolution than the final resolution of the reconstruction (Fig. 5A). The application of a low-pass filter prevents alignment of high-frequency noise. In this way, 3 reconstructions were generated from different tomograms collected at different defocuses. These reconstructions show higher-resolution features that are absent in the initial model. Fourier shell cross-correlation curves were calculated between different reconstructions. The curves show regions of negative correlation at higher resolutions, due to the different defocuses at which the tomograms were collected, and indicate that signal is present past the first node of the CTF (Fig. S3).
CTF correction was carried out as described in SI Text. Fourier shell cross-correlation curves between reconstructions from different reconstructions after CTF correction no longer show regions of negative correlation (Fig. S3).
The final reconstructions were aligned in 3D, averaged, and used as a starting model for a further round of subtomogram alignment. The 3 reconstructions were then averaged to generate a final reconstruction. The resolution of the reconstruction was measured as described in SI Text. Global lattice maps were visualized by placing a hexamer at the final aligned coordinates using the EM package for Amira (37); see also SI Text.
Supplementary Material
Acknowledgments.
We thank V. Vogt, A. Frangakis, and L.-A. Carlson for critical reading of the manuscript, and K. Grünewald and P. Prevelige for helpful comments. This work was supported by Deutsche Forschungsgemeinschaft grants within SPP1175 and SFB638 (to J.A.G.B. and H.-G.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903535106/DCSupplemental.
References
- 1.Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 3.Wilk T, et al. Organization of immature human immunodeficiency virus type 1. J Virol. 2001;75:759–771. doi: 10.1128/JVI.75.2.759-771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs JAG, Johnson MC, Simon MN, Fuller SD, Vogt VM. Cryo-electron microscopy reveals conserved and divergent features of Gag packing in immature particles of Rous Sarcoma virus and human immunodeficiency virus. J Mol Biol. 2006;355:157–168. doi: 10.1016/j.jmb.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 5.Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 7.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs JAG, et al. The mechanism of HIV-1 core assembly: Insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Briggs JAG, Wilk T, Welker R, Kräusslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Briggs JAG, et al. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 12.Fuller SD, Wilk T, Gowen BE, Kräusslich HG, Vogt VM. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 13.Wright ER, et al. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson LA, et al. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 18.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 19.Accola MA, Hoglund S, Gottlinger HG. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang C, et al. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J Virol. 2002;76:11729–11737. doi: 10.1128/JVI.76.22.11729-11737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morellet N, Druillennec S, Lenoir C, Bouaziz S, Roques BP. Helical structure determined by NMR of the HIV-1 (345–392)Gag sequence, surrounding p2: Implications for particle assembly and RNA packaging. Protein Sci. 2005;14:375–386. doi: 10.1110/ps.041087605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 23.Thali M, et al. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 24.Bartonova V, et al. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem. 2008;283:32024–32033. doi: 10.1074/jbc.M804230200. [DOI] [PubMed] [Google Scholar]
- 25.Gamble TR, et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 26.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 27.Ivanov D, et al. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci USA. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nature Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 29.Mortuza GB, et al. Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation. J Mol Biol. 2008;376:1493–1508. doi: 10.1016/j.jmb.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 30.Mortuza GB, et al. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 31.Cardone G, Purdy JG, Cheng N, Craven RC, Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dettenhofer M, Yu XF. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross I, Hohenberg H, Huckhagel C, Krausslich HG. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross I, et al. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruggnaller S, Mayr M, Frangakis AS. A visualization and segmentation toolbox for electron microscopy. J Struct Biol. 2008;164:161–165. doi: 10.1016/j.jsb.2008.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.