Figure 2. Glucose-6-phosphate dehydrogenase is activated by Src kinase in the liver of Zucker fa/fa rats.
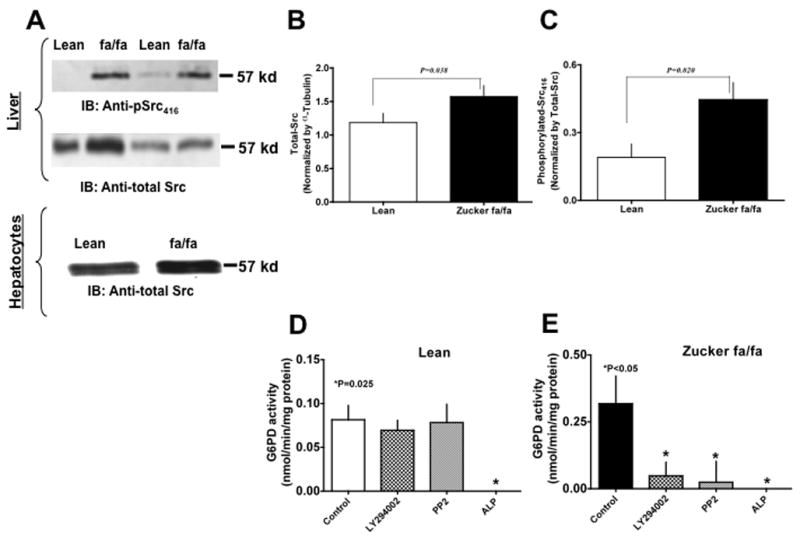
(A) Zucker lean (lanes 1 & 3) and fa/fa (lanes 2 & 4) rat liver homogenates (Panel A top) and hepatocyte lysates (Panel A bottom) were analyzed on 9% SDS-PAGE. Western blot analysis was performed using rabbit polyclonal anti-pSrc (first panel) and anti-total Src antibody (second panel). All input lanes contain 35 μg of total protein content. Panel (A) represents one blot of six such independent experiments. (B) & (C) Graphs represents total-Src (B) and phosphorylated-Src416 (C) in lean (n=6) and ZDF (n=6) rat liver as estimated by densitometric analysis and normalized with total-Src or α-tubulin values. (D) & (E) Lean (D) and Zucker fa/fa (E) liver tissues were incubated at 37°C for 10 minutes without and with PI3 kinase inhibitor, LY294002 (10 μM), Src kinase inhibitor, PP2 (10 μM), and alkaline phosphatase (200 U/ml), respectively. Graphs represents glucose-6-phosphate dehydrogenase activity in, Lean (D) and Zucker fa/fa (E) liver homogenates, respectively. Statistical analysis was performed using Student’s t-test and P<0.05 (lean versus Zucker fa/fa) was considered statistically significant.
