2.
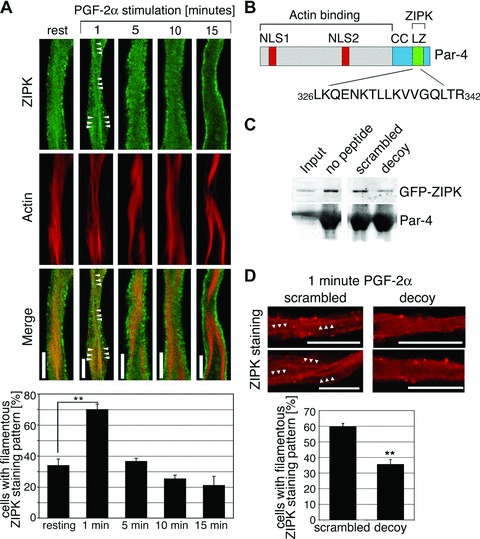
A Par-4 decoy peptide inhibits the transient association of ZIPK with actin filaments. (A) dVSMCs were stimulated with 10 μmol/L PGF-2α or left untreated. Cells were fixed and stained for ZIPK and actin. Scale bars, 5 μm. Graph: The percentage of cells with filamentous staining pattern (showing two or more filaments of at least 5 μm length) is plotted against different time-points of PGF-2α stimulation. Values are averages from three independent experiments with a total of 44–54 cells per time-point. (B) Amino acid sequence of the decoy peptide and its position within the Par-4 protein. NLS1 and NLS2, nuclear localization signals; CC, coiled coil region; LZ, leucine zipper motif. (C) Control experiment showing peptide specificity. Immobilized Par-4 protein was incubated with extracts from GFP-ZIPK transfected A7r5 cells in the absence or presence of the Par-4 decoy peptide or the scrambled control peptide. ZIPK binding to the recombinant Par-4 protein, as detected by western blotting, is reduced in the presence of the Par-4 decoy peptide. (D) dVSMCs were incubated with scrambled or decoy peptides prior to stimulation with 10 μmol/L PGF-2α for 1 min. Cells were fixed and stained for ZIPK. Two representative cells are shown. Arrowheads highlight filamentous structures. Graph: the percentage of cells with filamentous ZIPK staining pattern was assessed in microscopy experiments. The graph shows the statistical analysis of three independent experiments with a total of 51–74 cells per peptide. Scale bars, 10 μm (**P < 0.01).
