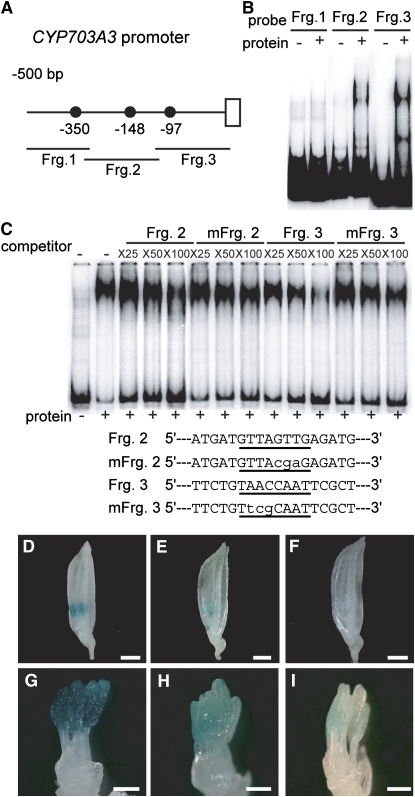
Figure 8.
In Vivo Function of GAMYB Binding-Like Motifs in CYP703A3.
(A) Schematic representation of the 5′-flanking region of CYP703A3. The closed circles show GAMYB binding-like motifs. Fragments 1, 2, and 3 (Frg. 1, bp −510 to −308; Frg. 2, bp −328 to −112; Frg. 3, bp −131 to +40) were used as probes or competitors for gel-shift assays in (B) and (C).
(B) Gel-shift assay with the recombinant GAMYB protein and 32P-labeled Frg. 1 to 3 presented in (A).
(C) Competitive gel-shift assay. The interaction between GAMYB and 32P-labeled RAmy1A probe was efficiently competed by Fragment 3 but not by Fragment 2, whereas mutagenized Fragment 2 or 3 (mFrg. 2 and 3), which contain displaced nucleotides at the GAMYB binding site, did not show effective competitive activity. Competition experiments were performed using increasing molar amounts (×25, ×50, and ×100) of the indicated unlabeled fragment. The sequences of the oligonucleotides used as competitor fragments are shown at the bottom of the panel. The putative binding site is underlined, and the mutations are indicated in lowercase letters.
(D) to (F) Whole flowers at the TD/YM stage. Bars = 1 mm.
(G) to (I) Close-up view of stamens at the TD/YM stage. Bars = 300 μm.
(D) and (G) Flower from a plant transformed with the intact ProCYP703A3:GUS construct (line #6).
(E) and (H) Flower from a plant transformed with the mutagenized ProCYP703A3 (mFrg. 2):GUS construct (line #12).
(F) and (I) Flower from a plant transformed with the mutagenized ProCYP703A3 construct (mFrg. 3):GUS (line #9). We analyzed 10 T0-independent lines for ProCYP703A3:GUS, 12 lines for mFrg. 2, and 18 lines for mFrg. 3.