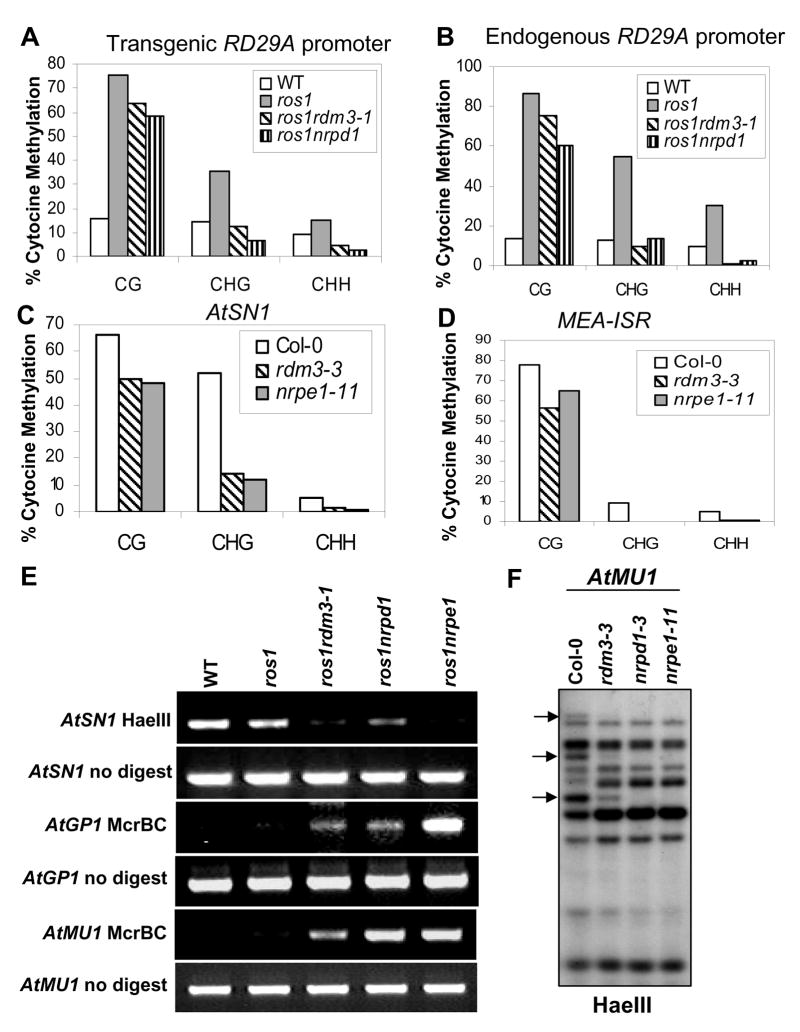
Figure 2. The rdm3 mutations reduce DNA methylation at RdDM target loci.
The percentage of cytosine methylation was determined by bisulfite sequencing at transgenic (A) and endogenous (B) RD29A promoters, AtSN1 (C) and MEA-ISR (D). The percentage of cytosine methylation on CG, CHG, and CHH sites is shown. H represents A, T, or C. (E) The rdm3-1 mutation suppressed DNA methylation in AtSN1, AtGP1, and AtMU1. After the indicated genomic DNA was digested with the methylation-sensitive restriction enzyme HaeIII, it was used for amplification of AtSN1. After the genomic DNA was digested with the methylated DNA-specific restriction enzyme McrBC, it was used for amplification of AtGP1 and AtMU1. The amplifications of non-digested genomic DNA were used as controls. (F) The rdm3-3 mutation reduced AtMU1 methylation at CHH sites. Genomic DNA from the indicated genotypes was digested with HaeIII, followed by Southern blot analysis. The three undigested bands (arrows) that are present in the Col-0 wild type were mostly digested in rdm3-3, nrpd1-3 and nrpe1-11.